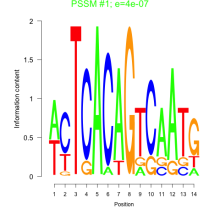
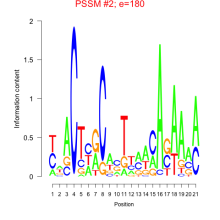
Phatr_bicluster_0233 Residual: 0.25
Phaeodactylum tricornutum
| Title | Residual | Model version | Organism |
|---|---|---|---|
| Phatr_bicluster_0233 | 0.25 | Phaeodactylum tricornutum |
Displaying 1 - 25 of 25
" class="views-fluidgrid-wrapper clear-block">
PHATRDRAFT_11021 branched-chain alpha-keto acid decarboxylase e1 beta subunit (PTZ00182)
PHATRDRAFT_1905 ibr domain protein (IBR)
PHATRDRAFT_20310 acyl-coenzyme ashort branched chain (ACAD superfamily)
PHATRDRAFT_35818 methylated-dna--protein-cysteine methyltransferase (ATase)
PHATRDRAFT_43899 riken cdna 2810055g22 gene
PHATRDRAFT_44106 amine oxidase (Amino_oxidase)
PHATRDRAFT_44412 riken cdnaisoform cra_b
PHATRDRAFT_45054 metallo-beta-lactamase family protein (GloB)
PHATRDRAFT_45160 (ANK)
PHATRDRAFT_45612 cog4371: membrane protein (DUF1517 superfamily)
PHATRDRAFT_46423 tumor differentially expressed 2-like (Serinc superfamily)
PHATRDRAFT_46752 (CHD)
PHATRDRAFT_47817 wd domain containingexpressed
PHATRDRAFT_54219 mgc85493 protein (PLN02528)
| GO ID | Go Term | p-value | q-value | Cluster |
|---|---|---|---|---|
| GO:0009190 | cyclic nucleotide biosynthesis | 0.0331273 | 1 | Phatr_bicluster_0233 |
| GO:0006281 | DNA repair | 0.0652904 | 1 | Phatr_bicluster_0233 |
| GO:0006118 | electron transport | 0.00808825 | 1 | Phatr_bicluster_0233 |
| GO:0007242 | intracellular signaling cascade | 0.0800992 | 1 | Phatr_bicluster_0233 |
| GO:0016567 | protein ubiquitination | 0.13382 | 1 | Phatr_bicluster_0233 |
| GO:0051341 | regulation of oxidoreductase activity | 0.0312056 | 1 | Phatr_bicluster_0233 |

Comments