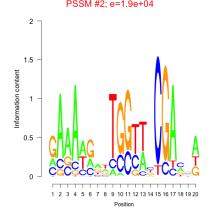
Phatr_bicluster_0234 Residual: 0.45
Phaeodactylum tricornutum
| Title | Residual | Model version | Organism |
|---|---|---|---|
| Phatr_bicluster_0234 | 0.45 | Phaeodactylum tricornutum |
Displaying 1 - 28 of 28
" class="views-fluidgrid-wrapper clear-block">
PHATRDRAFT_107 leucine-rich repeat transmembrane protein (LRR_RI superfamily)
PHATRDRAFT_11811 enoyl-hydratase isomerase (crotonase-like)
PHATRDRAFT_14327 nli interacting factorfamily protein (NIF)
PHATRDRAFT_19805 hypersensitive-induced response protein (SPFH_like_u4)
PHATRDRAFT_2761 purple acid phosphatase-like protein (MPP_superfamily superfamily)
PHATRDRAFT_33198 mesenchymal stem cell protein dscd75 (4HBT)
PHATRDRAFT_33901 atp binding protein serine threonine kinase (PLN00113)
PHATRDRAFT_34800 phosphoglycerate dehydrogenase-like protein (PLN02928)
PHATRDRAFT_36043 (DUF913 superfamily)
PHATRDRAFT_42500 (Band_7)
PHATRDRAFT_43674 (Beta_propel)
PHATRDRAFT_44326 (DUF563)
PHATRDRAFT_44442 nuclear localization sequence binding protein nsr1p (RRM)
PHATRDRAFT_45416 (WD40 superfamily)
PHATRDRAFT_46321 prenylated rab acceptor family protein (PRA1 superfamily)
PHATRDRAFT_49277 damaged dna binding protein (BTB)
PHATRDRAFT_49399 fis1_debha mitochondria fission 1 protein
PHATRDRAFT_49894 cyclin a-like protein
PHATRDRAFT_54319 secreted trypsin-like serine protease (Tryp_SPc)
PHATRDRAFT_54658 glue protein
PHATRDRAFT_54743 mgc83346 protein (WD40)
| GO ID | Go Term | p-value | q-value | Cluster |
|---|---|---|---|---|
| GO:0051028 | mRNA transport | 0.00172882 | 1 | Phatr_bicluster_0234 |
| GO:0006564 | L-serine biosynthesis | 0.013759 | 1 | Phatr_bicluster_0234 |
| GO:0000074 | regulation of cell cycle | 0.0573612 | 1 | Phatr_bicluster_0234 |
| GO:0006512 | ubiquitin cycle | 0.0639203 | 1 | Phatr_bicluster_0234 |
| GO:0008152 | metabolism | 0.0865285 | 1 | Phatr_bicluster_0234 |
| GO:0006118 | electron transport | 0.33492 | 1 | Phatr_bicluster_0234 |

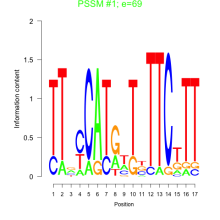
Comments