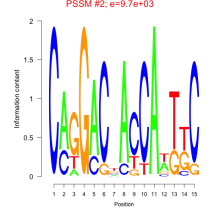
Phatr_bicluster_0236 Residual: 0.25
Phaeodactylum tricornutum
| Title | Residual | Model version | Organism |
|---|---|---|---|
| Phatr_bicluster_0236 | 0.25 | Phaeodactylum tricornutum |
Displaying 1 - 17 of 17
" class="views-fluidgrid-wrapper clear-block">
PHATRDRAFT_27246 dna2l protein (AAA_12)
PHATRDRAFT_39117 two component regulator three y domain protein
PHATRDRAFT_43317 (DUF2088 superfamily)
PHATRDRAFT_44222 enhanced disease susceptibility 5 (MATE_like superfamily)
PHATRDRAFT_44868 leucine aminopeptidase (PRK00913)
PHATRDRAFT_47791 (Ecm29 superfamily)
PHATRDRAFT_47881 af225976_1 flavoredoxin (Flavin_Reduct)
PHATRDRAFT_48187 protein familywith a coiled coil-4 domain (Methyltransf_22)
PHATRDRAFT_49951 trans-golgi p230
PHATRDRAFT_50146 amino acid transporter (Aa_trans superfamily)
PHATRDRAFT_50470 (zf-MYND)
PHATRDRAFT_52678 (POLBc_epsilon)
| GO ID | Go Term | p-value | q-value | Cluster |
|---|---|---|---|---|
| GO:0006865 | amino acid transport | 0.0206078 | 1 | Phatr_bicluster_0236 |
| GO:0019538 | protein metabolism | 0.0264433 | 1 | Phatr_bicluster_0236 |
| GO:0006508 | proteolysis and peptidolysis | 0.156991 | 1 | Phatr_bicluster_0236 |


Comments