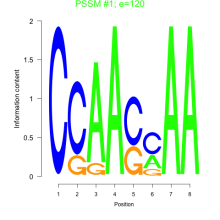
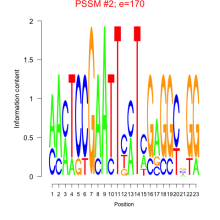
Phatr_bicluster_0237 Residual: 0.20
Phaeodactylum tricornutum
| Title | Residual | Model version | Organism |
|---|---|---|---|
| Phatr_bicluster_0237 | 0.20 | Phaeodactylum tricornutum |
Displaying 1 - 21 of 21
" class="views-fluidgrid-wrapper clear-block">
PHATRDRAFT_12144 chromosome 1 open reading frame 33 (Ribosomal_P0_like)
PHATRDRAFT_19647 brix domain containing 1 (Brix superfamily)
PHATRDRAFT_20740 rna binding motif protein 19 (RRM3_RBM19_RRM2_MRD1)
PHATRDRAFT_25133 nog2_klula nucleolar gtp-binding protein 2 (NGP_1)
PHATRDRAFT_25549 dead (asp-glu-ala-asp) box polypeptide 56 (DEXDc superfamily)
PHATRDRAFT_26035 (Ribosomal_L1)
PHATRDRAFT_27905 peter pan homolog (Brix)
PHATRDRAFT_31725 (PIN_Nob1)
PHATRDRAFT_32758 dead box rna helicase (PRK04837)
PHATRDRAFT_32836 (RRM_Nop6)
PHATRDRAFT_42570 ribosomal rna processing 12 homolog (NUC173 superfamily)
PHATRDRAFT_43574 transducin family protein wd-40 repeat family protein (WD40 superfamily)
PHATRDRAFT_44555 zz domain protein (Kri1_C)
PHATRDRAFT_45888 nucleolar complex associated 4 homolog (CBF)
PHATRDRAFT_46846 glioma tumor suppressor candidate region gene 2 isoform 4 (Nop53)
PHATRDRAFT_47058 (Pumilio superfamily)
PHATRDRAFT_49441 helicase domain protein (HA)
PHATRDRAFT_51007 u3 small nucleolar ribonucleoprotein protein (Brix)
| GO ID | Go Term | p-value | q-value | Cluster |
|---|---|---|---|---|
| GO:0016071 | mRNA metabolism | 0.0029615 | 1 | Phatr_bicluster_0237 |
| GO:0006412 | protein biosynthesis | 0.114635 | 1 | Phatr_bicluster_0237 |
| GO:0042254 | ribosome biogenesis and assembly | 0.00444005 | 1 | Phatr_bicluster_0237 |

Comments