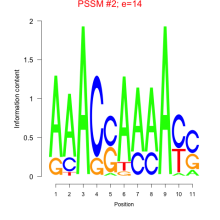
Phatr_bicluster_0246 Residual: 0.27
Phaeodactylum tricornutum
| Title | Residual | Model version | Organism |
|---|---|---|---|
| Phatr_bicluster_0246 | 0.27 | Phaeodactylum tricornutum |
Displaying 1 - 25 of 25
" class="views-fluidgrid-wrapper clear-block">
PHATRDRAFT_11458 26s proteasome regulatory subunit (MPN_eIF3f)
PHATRDRAFT_11850 elac homolog 1 (Lactamase_B superfamily)
PHATRDRAFT_12238 50s ribosomal protein l4 (Ribosomal_L4 superfamily)
PHATRDRAFT_13088 eukaryotic translation initiation factorsubunit 7 (eIF-3_zeta)
PHATRDRAFT_17545 cyc07-like protein (Ribosomal_S3Ae)
PHATRDRAFT_18246 5-enolpyruvylshikimate-3-phosphate synthase (PLN02338)
PHATRDRAFT_21181 quinoid dihydropteridine reductase (NADB_Rossmann superfamily)
PHATRDRAFT_21485 eukaryotic translation initiation factorsubunit 6 (eIF3_N)
PHATRDRAFT_21592 diaminopimelate decarboxylase (PLPDE_III_DapDC)
PHATRDRAFT_22525 ubiquitin conjugating enzyme (UBCc)
PHATRDRAFT_22666 chaperonin subunit 6a (TCP1_zeta)
PHATRDRAFT_23371 gtp-binding protein (odn superfamily) (PTZ00258)
PHATRDRAFT_27385 glutamine-trna ligase (PRK05347)
PHATRDRAFT_27838 elongation factor 3 (Uup)
PHATRDRAFT_35513 prpl12 chloroplast ribosomal protein l12 (Ribosomal_L7_L12)
PHATRDRAFT_37131 at3g11830 f26k24_12 (TCP1_eta)
PHATRDRAFT_40621 gtp-binding protein (TypA)
PHATRDRAFT_43732 (CLU)
PHATRDRAFT_46847 riken cdnaisoform cra_b (CTD_bind)
PHATRDRAFT_47027 proteasome family protein (PCI)
PHATRDRAFT_48822 translation elongation factor (GST_C_eEF1b_like)
PHATRDRAFT_54376 (DnaJ)
| GO ID | Go Term | p-value | q-value | Cluster |
|---|---|---|---|---|
| GO:0044267 | cellular protein metabolism | 0.000646869 | 1 | Phatr_bicluster_0246 |
| GO:0006412 | protein biosynthesis | 0.00301306 | 1 | Phatr_bicluster_0246 |
| GO:0016089 | aromatic amino acid family biosynthesis, shikimate pathway | 0.00395159 | 1 | Phatr_bicluster_0246 |
| GO:0006424 | glutamyl-tRNA aminoacylation | 0.00788854 | 1 | Phatr_bicluster_0246 |
| GO:0009089 | lysine biosynthesis via diaminopimelate | 0.0118109 | 1 | Phatr_bicluster_0246 |
| GO:0007186 | G-protein coupled receptor protein signaling pathway | 0.0502429 | 1 | Phatr_bicluster_0246 |
| GO:0006414 | translational elongation | 0.0540081 | 1 | Phatr_bicluster_0246 |
| GO:0006413 | translational initiation | 0.0908976 | 1 | Phatr_bicluster_0246 |
| GO:0006512 | ubiquitin cycle | 0.140282 | 1 | Phatr_bicluster_0246 |
| GO:0006418 | tRNA aminoacylation for protein translation | 0.187143 | 1 | Phatr_bicluster_0246 |

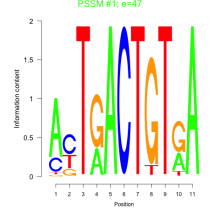
Comments