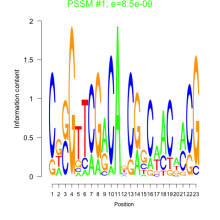
Phatr_bicluster_0247 Residual: 0.46
Phaeodactylum tricornutum
| Title | Residual | Model version | Organism |
|---|---|---|---|
| Phatr_bicluster_0247 | 0.46 | Phaeodactylum tricornutum |
Displaying 1 - 29 of 29
" class="views-fluidgrid-wrapper clear-block">
PHATRDRAFT_11401 elicitin protein ram6
PHATRDRAFT_33124 copia protein (gag-int-pol protein)
PHATRDRAFT_34085 (Glyoxalase_3)
PHATRDRAFT_37580 mucin 2 precursor
PHATRDRAFT_37902 mucin 2 precursor
PHATRDRAFT_38388 mucin 2 precursor
PHATRDRAFT_39762 mucin 2 precursor
PHATRDRAFT_39794 heat shock factor cg5748-isoform d (HSF_DNA-bind superfamily)
PHATRDRAFT_40843 mucin 2 precursor
PHATRDRAFT_42322 casein kinase (Pkinase)
PHATRDRAFT_45384 (DnaJ)
PHATRDRAFT_54547 casein kinase (Pkinase)
PHATRDRAFT_8537 serine threonine kinase with two-component sensor domain (COG3899)
PHATRDRAFT_8975 (kgd)
| GO ID | Go Term | p-value | q-value | Cluster |
|---|---|---|---|---|
| GO:0006310 | DNA recombination | 0.00000398 | 0.00922066 | Phatr_bicluster_0247 |
| GO:0046168 | glycerol-3-phosphate catabolism | 0.00641182 | 1 | Phatr_bicluster_0247 |
| GO:0006072 | glycerol-3-phosphate metabolism | 0.0127857 | 1 | Phatr_bicluster_0247 |
| GO:0007018 | microtubule-based movement | 0.0922831 | 1 | Phatr_bicluster_0247 |
| GO:0006468 | protein amino acid phosphorylation | 0.102976 | 1 | Phatr_bicluster_0247 |
| GO:0007165 | signal transduction | 0.141021 | 1 | Phatr_bicluster_0247 |
| GO:0005975 | carbohydrate metabolism | 0.184674 | 1 | Phatr_bicluster_0247 |
| GO:0006457 | protein folding | 0.317339 | 1 | Phatr_bicluster_0247 |
| GO:0006355 | regulation of transcription, DNA-dependent | 0.515177 | 1 | Phatr_bicluster_0247 |

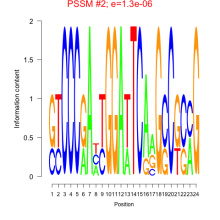
Comments