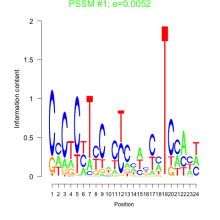
Thaps_bicluster_0004 Residual: 0.42
Thalassiosira pseudonana
| Title | Residual | Model version | Organism |
|---|---|---|---|
| Thaps_bicluster_0004 | 0.42 | Thalassiosira pseudonana |
Displaying 1 - 39 of 39
" class="views-fluidgrid-wrapper clear-block">
11093 GH99_GH71_like_2
11242 hypothetical protein
19553 PX_domain superfamily
2070 hypothetical protein
22142 hypothetical protein
22382 hypothetical protein
23108 hypothetical protein
24452 hypothetical protein
25444 Glyco_transf_22 superfamily
25734 hypothetical protein
260785 NOX_Duox_like_FAD_NADP
262620 ANK
262645 Glyco_18
268368 (Tp_HSF_3.6a) HSF_DNA-bind superfamily
269718 sucA
269868 MviM
30335 hypothetical protein
3052 hypothetical protein
31298 S_TKc
3137 hypothetical protein
3394 hypothetical protein
34186 SPC12
35281 PKc_like superfamily
35519 hypothetical protein
37959 hypothetical protein
38335 (PSB3) proteasome_beta_type_3
39955 ARM
4043 KAP
5301 hypothetical protein
5801 hypothetical protein
6982 hypothetical protein
7623 hypothetical protein
8330 hypothetical protein
8518 hypothetical protein
8615 hypothetical protein
9475 hypothetical protein
953 hypothetical protein
9953 hypothetical protein
9998 BTB superfamily
| GO ID | Go Term | p-value | q-value | Cluster |
|---|---|---|---|---|
| GO:0015986 | ATP synthesis coupled proton transport | 0.133715 | 1 | Thaps_bicluster_0004 |
| GO:0005975 | carbohydrate metabolism | 0.209229 | 1 | Thaps_bicluster_0004 |
| GO:0006118 | electron transport | 0.198115 | 1 | Thaps_bicluster_0004 |
| GO:0007242 | intracellular signaling cascade | 0.122787 | 1 | Thaps_bicluster_0004 |
| GO:0008152 | metabolism | 0.126308 | 1 | Thaps_bicluster_0004 |
| GO:0009401 | phosphoenolpyruvate-dependent sugar phosphotransferase system | 0.0544657 | 1 | Thaps_bicluster_0004 |
| GO:0006813 | potassium ion transport | 0.066187 | 1 | Thaps_bicluster_0004 |
| GO:0006468 | protein amino acid phosphorylation | 0.139278 | 1 | Thaps_bicluster_0004 |
| GO:0006355 | regulation of transcription, DNA-dependent | 0.626585 | 1 | Thaps_bicluster_0004 |
| GO:0006465 | signal peptide processing | 0.0123506 | 1 | Thaps_bicluster_0004 |


Comments