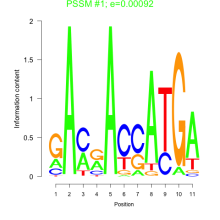
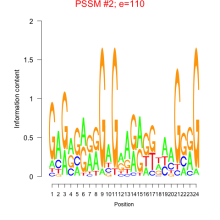
Thaps_bicluster_0006 Residual: 0.41
Thalassiosira pseudonana
| Title | Residual | Model version | Organism |
|---|---|---|---|
| Thaps_bicluster_0006 | 0.41 | Thalassiosira pseudonana |
Displaying 1 - 53 of 53
" class="views-fluidgrid-wrapper clear-block">
1023 hypothetical protein
1031 hypothetical protein
11118 EFh
11346 hypothetical protein
11623 HSF_DNA-bind superfamily
1204 hypothetical protein
12128 hypothetical protein
18158 DUF2453
18665 (CLPP2) S14_ClpP_2
1870 COG3899
22656 metallo-dependent_hydrolases superfamily
22810 hypothetical protein
23505 hypothetical protein
24087 hypothetical protein
25041 hypothetical protein
25369 DUF4517 superfamily
25833 hypothetical protein
261492 HAM1
261657 ZntA
262296 Hydrolase_4 superfamily
263354 Fer4_NifH superfamily
263682 hypothetical protein
264717 Peptidase_M14_like superfamily
268988 (CPF3) crypto_DASH
270141 (KAS3) PLN02326
3022 hypothetical protein
3065 hypothetical protein
3160 hypothetical protein
32319 5_nucleotid superfamily
3256 hypothetical protein
33973 Patatin_and_cPLA2 superfamily
3587 SRPBCC superfamily
37944 UPF0061 superfamily
37961 tim10-like protein
39936 MPP_PP1_PPKL
4215 LRR_RI superfamily
4405 hypothetical protein
4758 (Tp_HSF_2.2e) HSF_DNA-bind superfamily
5265 VEFS-Box superfamily
5328 hypothetical protein
5657 hypothetical protein
6240 hypothetical protein
7094 COG1233
7330 NYN_YacP
7462 hypothetical protein
8227 hypothetical protein
8449 hypothetical protein
8740 hypothetical protein
943 hypothetical protein
9710 (mapk) PKc_like superfamily
9737 hypothetical protein
9756 Ribosomal_L21p superfamily
ThpsCp085 (psbA) photosystem II protein D1
| GO ID | Go Term | p-value | q-value | Cluster |
|---|---|---|---|---|
| GO:0045039 | mitochondrial inner membrane protein import | 0.0156247 | 1 | Thaps_bicluster_0006 |
| GO:0006626 | protein-mitochondrial targeting | 0.0156247 | 1 | Thaps_bicluster_0006 |
| GO:0006461 | protein complex assembly | 0.0271911 | 1 | Thaps_bicluster_0006 |
| GO:0016117 | carotenoid biosynthesis | 0.0310179 | 1 | Thaps_bicluster_0006 |
| GO:0030001 | metal ion transport | 0.0499389 | 1 | Thaps_bicluster_0006 |
| GO:0006633 | fatty acid biosynthesis | 0.0721821 | 1 | Thaps_bicluster_0006 |
| GO:0006812 | cation transport | 0.156318 | 1 | Thaps_bicluster_0006 |
| GO:0015986 | ATP synthesis coupled proton transport | 0.166312 | 1 | Thaps_bicluster_0006 |
| GO:0006281 | DNA repair | 0.166312 | 1 | Thaps_bicluster_0006 |
| GO:0006418 | tRNA aminoacylation for protein translation | 0.202004 | 1 | Thaps_bicluster_0006 |

Comments