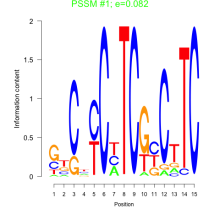
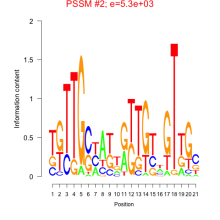
Thaps_bicluster_0022 Residual: 0.44
Thalassiosira pseudonana
| Title | Residual | Model version | Organism |
|---|---|---|---|
| Thaps_bicluster_0022 | 0.44 | Thalassiosira pseudonana |
Displaying 1 - 50 of 50
" class="views-fluidgrid-wrapper clear-block">
11312 hypothetical protein
11487 PhyH superfamily
12205 hypothetical protein
14638 Pkinase
1586 UreD superfamily
19360 HELICc
19899 Bromodomain superfamily
20639 hypothetical protein
22339 U-box superfamily
22351 Zip superfamily
22758 Dynein_light superfamily
22859 hypothetical protein
24305 Ubox
24762 hypothetical protein
25635 hypothetical protein
25651 hypothetical protein
25794 hypothetical protein
25879 TopA
262186 KISc
262585 NAD_binding_8
262815 (Tp_Myb1R_SHAQKYF2) MYB DNA binding protein/ transcription factor-like protein
263138 CysPc superfamily
263583 ankyrin domain-containing protein
263978 (Tp_HSF_4.2b) HSF_DNA-bind
264346 (Tp_HSF_3.7b) HSF_DNA-bind
264573 hypothetical protein
268311 hypothetical protein
268493 (KAR1) NADB_Rossmann superfamily
30950 Glyco_hydro_47
31035 Trypsin_2
31046 LbH_gamma_CA_like
3131 S_TKc
32522 rad3
35984 hypothetical protein
36249 hypothetical protein
3635 hypothetical protein
37562 TyrKc
4061 hypothetical protein
4263 hypothetical protein
5405 hypothetical protein
5774 (Tp_HSF_3.5e) regulator [Rayko]
6126 hypothetical protein
6745 hypothetical protein
7180 hypothetical protein
7370 hypothetical protein
7648 MIP-T3
9142 hypothetical protein
9667 hypothetical protein
9678 hypothetical protein
ThpsCp091 (thiS) thiamine biosynthesis protein
| GO ID | Go Term | p-value | q-value | Cluster |
|---|---|---|---|---|
| GO:0016567 | protein ubiquitination | 0.00662963 | 1 | Thaps_bicluster_0022 |
| GO:0007017 | microtubule-based process | 0.0148146 | 1 | Thaps_bicluster_0022 |
| GO:0000103 | sulfate assimilation | 0.0197059 | 1 | Thaps_bicluster_0022 |
| GO:0006268 | DNA unwinding | 0.0245739 | 1 | Thaps_bicluster_0022 |
| GO:0006487 | N-linked glycosylation | 0.0245739 | 1 | Thaps_bicluster_0022 |
| GO:0006289 | nucleotide-excision repair | 0.0245739 | 1 | Thaps_bicluster_0022 |
| GO:0030001 | metal ion transport | 0.0626925 | 1 | Thaps_bicluster_0022 |
| GO:0006139 | nucleobase, nucleoside, nucleotide and nucleic acid metabolism | 0.0719967 | 1 | Thaps_bicluster_0022 |
| GO:0006468 | protein amino acid phosphorylation | 0.0863768 | 1 | Thaps_bicluster_0022 |
| GO:0006306 | DNA methylation | 0.134692 | 1 | Thaps_bicluster_0022 |

Comments