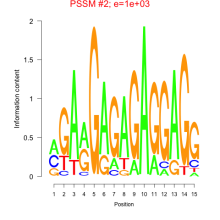
Thaps_bicluster_0030 Residual: 0.47
Thalassiosira pseudonana
| Title | Residual | Model version | Organism |
|---|---|---|---|
| Thaps_bicluster_0030 | 0.47 | Thalassiosira pseudonana |
Displaying 1 - 45 of 45
" class="views-fluidgrid-wrapper clear-block">
10903 hypothetical protein
10927 hypothetical protein
11129 hypothetical protein
11543 hypothetical protein
11548 MDR8
19913 ybbN
20821 SCP superfamily
21592 hypothetical protein
21900 COG1233
22681 hypothetical protein
23271 hypothetical protein
24067 hypothetical protein
24383 hypothetical protein
24435 2a58
24564 hypothetical protein
24726 PLN00113
2496 hypothetical protein
25088 hypothetical protein
25439 hypothetical protein
263081 PLN00113
26365 fadJ
264577 lysozyme_like superfamily
268234 hypothetical protein
269307 hypothetical protein
269316 fadE
270137 EFh
3075 hypothetical protein
30804 AdoMet_dc
31062 WD40
3108 hypothetical protein
31722 HMGB-UBF_HMG-box
34809 (KCT1) PRK05790
35710 (ACD1) SCAD_SBCAD
3627 fadI
38047 RCC1
38231 (PETC2) PRK13474
38907 Abhydrolase_5
4348 hypothetical protein
6422 hypothetical protein
7974 hypothetical protein
9145 hypothetical protein
9240 hypothetical protein
9500 hypothetical protein
9552 hypothetical protein
ThpsCr004 (rrn5')
| GO ID | Go Term | p-value | q-value | Cluster |
|---|---|---|---|---|
| GO:0006118 | electron transport | 0.00311497 | 1 | Thaps_bicluster_0030 |
| GO:0006631 | fatty acid metabolism | 0.00742571 | 1 | Thaps_bicluster_0030 |
| GO:0006817 | phosphate transport | 0.0123481 | 1 | Thaps_bicluster_0030 |
| GO:0016117 | carotenoid biosynthesis | 0.0196897 | 1 | Thaps_bicluster_0030 |
| GO:0006032 | chitin catabolism | 0.0196897 | 1 | Thaps_bicluster_0030 |
| GO:0016998 | cell wall catabolism | 0.0221257 | 1 | Thaps_bicluster_0030 |
| GO:0009613 | response to pest, pathogen or parasite | 0.0221257 | 1 | Thaps_bicluster_0030 |
| GO:0006355 | regulation of transcription, DNA-dependent | 0.545155 | 1 | Thaps_bicluster_0030 |
| GO:0008152 | metabolism | 0.652155 | 1 | Thaps_bicluster_0030 |

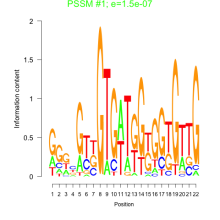
Comments