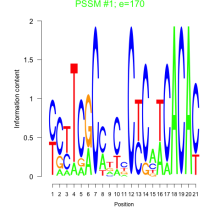
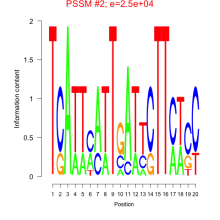
Thaps_bicluster_0043 Residual: 0.36
Thalassiosira pseudonana
| Title | Residual | Model version | Organism |
|---|---|---|---|
| Thaps_bicluster_0043 | 0.36 | Thalassiosira pseudonana |
Displaying 1 - 32 of 32
" class="views-fluidgrid-wrapper clear-block">
10106 hypothetical protein
10109 hypothetical protein
10750 MDR8
10986 hypothetical protein
11231 hypothetical protein
11377 hypothetical protein
11641 GluZincin superfamily
1455 hypothetical protein
1957 hypothetical protein
21138 AAA_34 superfamily
21370 ATS1
23434 DOMON_DOH
2613 hypothetical protein
261541 RRM3_CELF1-6
264486 integrin domain-containing protein
264853 (Tp_Myb3R1) SANT
35404 PTZ00018
4166 hypothetical protein
4491 hypothetical protein
4879 hypothetical protein
5192 hypothetical protein
5425 hypothetical protein
6541 hypothetical protein
6626 hypothetical protein
7122 (sig2) sig2-like protein
7307 hypothetical protein
7594 hypothetical protein
8203 hypothetical protein
8215 (Tp_HSF_3.5h) HSF_DNA-bind superfamily
8263 hypothetical protein
8511 hypothetical protein
9555 hypothetical protein
| GO ID | Go Term | p-value | q-value | Cluster |
|---|---|---|---|---|
| GO:0030163 | protein catabolic process | 0.00454357 | 1 | Thaps_bicluster_0043 |
| GO:0006508 | proteolysis | 0.0314132 | 1 | Thaps_bicluster_0043 |
| GO:0007160 | cell-matrix adhesion | 0.0402221 | 1 | Thaps_bicluster_0043 |
| GO:0007186 | G-protein coupled receptor protein signaling pathway | 0.0467809 | 1 | Thaps_bicluster_0043 |
| GO:0007001 | chromosome organization and biogenesis (sensu Eukaryota) | 0.0640724 | 1 | Thaps_bicluster_0043 |
| GO:0007155 | cell adhesion | 0.0662137 | 1 | Thaps_bicluster_0043 |
| GO:0006334 | nucleosome assembly | 0.0683505 | 1 | Thaps_bicluster_0043 |
| GO:0006355 | regulation of transcription, DNA-dependent | 0.514255 | 1 | Thaps_bicluster_0043 |

Comments