Thaps_bicluster_0051 Residual: 0.39
Thalassiosira pseudonana
| Title | Residual | Model version | Organism |
|---|---|---|---|
| Thaps_bicluster_0051 | 0.39 | Thalassiosira pseudonana |
Displaying 1 - 37 of 37
" class="views-fluidgrid-wrapper clear-block">
10279 hypothetical protein
10571 hypothetical protein
11719 hypothetical protein
12120 NADB_Rossmann superfamily
1397 hypothetical protein
17484 Nudix_Hydrolase superfamily
18771 MORN superfamily
20810 hypothetical protein
20987 hypothetical protein
21789 hypothetical protein
22673 NA
23607 hypothetical protein
24603 (Tp_HSF_2.6b) HSF_DNA-bind superfamily
24758 hypothetical protein
24864 hypothetical protein
24970 hypothetical protein
25953 (MDH2) PLN00105
260942 YjgF_YER057c_UK114_like_1
261112 Nudix_Hydrolase superfamily
263365 PP2Cc superfamily
264109 fabG
268009 Ftr1_plasma membrane iron permease
2705 COG2363
3064 hypothetical protein
3142 Mito_carr
3153 hypothetical protein
3302 hypothetical protein
3384 hypothetical protein
5502 hypothetical protein
5548 hypothetical protein
6595 hypothetical protein
687 PTZ00318
7678 hypothetical protein
7956 Cupin_3
874 hypothetical protein
9742 HA
9987 hypothetical protein
| GO ID | Go Term | p-value | q-value | Cluster |
|---|---|---|---|---|
| GO:0008299 | isoprenoid biosynthesis | 0.0180619 | 1 | Thaps_bicluster_0051 |
| GO:0006355 | regulation of transcription, DNA-dependent | 0.0872056 | 1 | Thaps_bicluster_0051 |
| GO:0008152 | metabolism | 0.141031 | 1 | Thaps_bicluster_0051 |
| GO:0006412 | protein biosynthesis | 0.243897 | 1 | Thaps_bicluster_0051 |
| GO:0006810 | transport | 0.266912 | 1 | Thaps_bicluster_0051 |
| GO:0006118 | electron transport | 0.364179 | 1 | Thaps_bicluster_0051 |


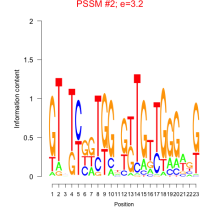
Comments