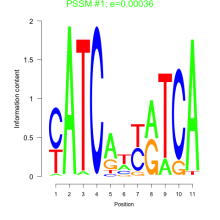
Thaps_bicluster_0057 Residual: 0.46
Thalassiosira pseudonana
| Title | Residual | Model version | Organism |
|---|---|---|---|
| Thaps_bicluster_0057 | 0.46 | Thalassiosira pseudonana |
Displaying 1 - 37 of 37
" class="views-fluidgrid-wrapper clear-block">
10073 hypothetical protein
10722 Lipase superfamily
10935 Ion_trans
10983 hypothetical protein
12135 hypothetical protein
1230 hypothetical protein
1243 hypothetical protein
1436 hypothetical protein
1755 hypothetical protein
21058 hypothetical protein
21583 hypothetical protein
21977 hypothetical protein
22639 hypothetical protein
23225 hypothetical protein
23755 hypothetical protein
24466 hypothetical protein
24535 MDR8
24636 hypothetical protein
2530 hypothetical protein
260761 HA
263035 HP_PGM_like
268362 (Tp_AP2-EREBP2) regulator [Rayko]
268958 hypothetical protein
34855 Abhydrolase_5
37123 (VAT2) V-ATPase_V1_A
37592 S_TKc
4095 hypothetical protein
4167 hypothetical protein
4659 hypothetical protein
4800 hypothetical protein
4946 hypothetical protein
5479 hypothetical protein
7895 hypothetical protein
8232 hypothetical protein
9092 hypothetical protein
9221 hypothetical protein
9558 hypothetical protein
| GO ID | Go Term | p-value | q-value | Cluster |
|---|---|---|---|---|
| GO:0006754 | ATP biosynthesis | 0.0123506 | 1 | Thaps_bicluster_0057 |
| GO:0006412 | translation | 0.0426071 | 1 | Thaps_bicluster_0057 |
| GO:0006811 | ion transport | 0.0544657 | 1 | Thaps_bicluster_0057 |
| GO:0006813 | potassium ion transport | 0.066187 | 1 | Thaps_bicluster_0057 |
| GO:0006629 | lipid metabolism | 0.0863737 | 1 | Thaps_bicluster_0057 |
| GO:0006096 | glycolysis | 0.108945 | 1 | Thaps_bicluster_0057 |
| GO:0006812 | cation transport | 0.125531 | 1 | Thaps_bicluster_0057 |
| GO:0015986 | ATP synthesis coupled proton transport | 0.133715 | 1 | Thaps_bicluster_0057 |
| GO:0006418 | tRNA aminoacylation for protein translation | 0.163117 | 1 | Thaps_bicluster_0057 |
| GO:0016567 | protein ubiquitination | 0.219149 | 1 | Thaps_bicluster_0057 |


Comments