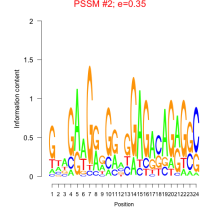
Thaps_bicluster_0067 Residual: 0.47
Thalassiosira pseudonana
| Title | Residual | Model version | Organism |
|---|---|---|---|
| Thaps_bicluster_0067 | 0.47 | Thalassiosira pseudonana |
Displaying 1 - 25 of 25
" class="views-fluidgrid-wrapper clear-block">
10143 hypothetical protein
11214 SOUL
1599 hypothetical protein
17673 DUF4033
1884 hypothetical protein
1937 hypothetical protein
20784 hypothetical protein
22494 COG0384
22703 hypothetical protein
24549 hypothetical protein
262637 P4Hc
263660 (LAT1_1) LPLAT_AGPAT-like
264603 SEC14
270376 hypothetical protein
27048 TM_PBP1_branched-chain-AA_like superfamily
28125 PLN00203
30924 NAD_binding_5 superfamily
3186 hypothetical protein
35464 Nth
37725 Coq4 superfamily
4710 KefB
5959 hypothetical protein
8329 GH_J
8409 hypothetical protein
9436 2OG-FeII_Oxy_3
| GO ID | Go Term | p-value | q-value | Cluster |
|---|---|---|---|---|
| GO:0019538 | protein metabolism | 0.00238909 | 1 | Thaps_bicluster_0067 |
| GO:0006021 | myo-inositol biosynthesis | 0.00330544 | 1 | Thaps_bicluster_0067 |
| GO:0006284 | base-excision repair | 0.00988766 | 1 | Thaps_bicluster_0067 |
| GO:0006779 | porphyrin biosynthesis | 0.0180619 | 1 | Thaps_bicluster_0067 |
| GO:0008654 | phospholipid biosynthesis | 0.0229381 | 1 | Thaps_bicluster_0067 |
| GO:0009058 | biosynthesis | 0.0874873 | 1 | Thaps_bicluster_0067 |
| GO:0008152 | metabolism | 0.505245 | 1 | Thaps_bicluster_0067 |

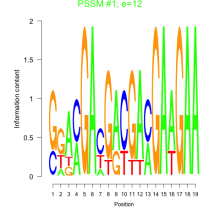
Comments