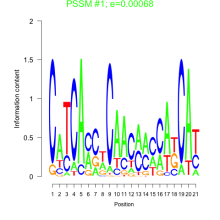
Thaps_bicluster_0070 Residual: 0.44
Thalassiosira pseudonana
| Title | Residual | Model version | Organism |
|---|---|---|---|
| Thaps_bicluster_0070 | 0.44 | Thalassiosira pseudonana |
Displaying 1 - 30 of 30
" class="views-fluidgrid-wrapper clear-block">
10833 hypothetical protein
11219 hypothetical protein
12063 hypothetical protein
13149 (DIC2) WD40 superfamily
1477 hypothetical protein
1555 hypothetical protein
1783 Meckelin superfamily
21387 Peptidase_M48 superfamily
22031 Sin_N superfamily
2217 hypothetical protein
2219 hypothetical protein
22354 hypothetical protein
22508 hypothetical protein
2321 DnaQ_like_exo superfamily
260941 CDP-OH_P_transf superfamily
261481 FTO_NTD
263499 CdsA
269065 Methyltransf_4 superfamily
269333 PTR2 superfamily
270310 hypothetical protein
3190 hypothetical protein
36506 PseudoU_synth superfamily
37559 DUF572
38289 KefB
38735 hypothetical protein
38770 AAA
3970 hypothetical protein
41246 PRK14333
5789 hypothetical protein
6638 ANK
| GO ID | Go Term | p-value | q-value | Cluster |
|---|---|---|---|---|
| GO:0008654 | phospholipid biosynthesis | 0.0172498 | 1 | Thaps_bicluster_0070 |
| GO:0008033 | tRNA processing | 0.0221325 | 1 | Thaps_bicluster_0070 |
| GO:0007186 | G-protein coupled receptor protein signaling pathway | 0.0257813 | 1 | Thaps_bicluster_0070 |
| GO:0006813 | potassium ion transport | 0.026995 | 1 | Thaps_bicluster_0070 |
| GO:0006508 | proteolysis and peptidolysis | 0.308243 | 1 | Thaps_bicluster_0070 |
| GO:0006355 | regulation of transcription, DNA-dependent | 0.325408 | 1 | Thaps_bicluster_0070 |


Comments