Thaps_bicluster_0071 Residual: 0.35
Thalassiosira pseudonana
| Title | Residual | Model version | Organism |
|---|---|---|---|
| Thaps_bicluster_0071 | 0.35 | Thalassiosira pseudonana |
Displaying 1 - 22 of 22
" class="views-fluidgrid-wrapper clear-block">
1291 hypothetical protein
16737 Mito_carr
21591 HtpX
21887 hypothetical protein
22682 hypothetical protein
22778 hypothetical protein
263006 P4Hc
263007 hypothetical protein
263061 Sod_Fe_N superfamily
263800 Zip superfamily
270296 hypothetical protein
2944 hypothetical protein
30380 (TPI2) TIM
35025 Lung_7-TM_R superfamily
3529 DnaQ_like_exo superfamily
4114 GAT_SF
4460 hypothetical protein
4997 hypothetical protein
5807 hypothetical protein
6066 APG6
8514 hypothetical protein
8862 hypothetical protein
| GO ID | Go Term | p-value | q-value | Cluster |
|---|---|---|---|---|
| GO:0006801 | superoxide metabolism | 0.00495406 | 1 | Thaps_bicluster_0071 |
| GO:0030001 | metal ion transport | 0.016026 | 1 | Thaps_bicluster_0071 |
| GO:0019538 | protein metabolism | 0.0557488 | 1 | Thaps_bicluster_0071 |
| GO:0006810 | transport | 0.207702 | 1 | Thaps_bicluster_0071 |
| GO:0006508 | proteolysis and peptidolysis | 0.308243 | 1 | Thaps_bicluster_0071 |
| GO:0008152 | metabolism | 0.410014 | 1 | Thaps_bicluster_0071 |

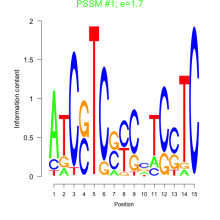
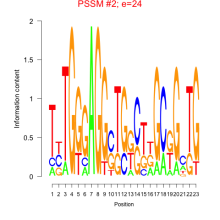
Comments