Thaps_bicluster_0072 Residual: 0.37
Thalassiosira pseudonana
| Title | Residual | Model version | Organism |
|---|---|---|---|
| Thaps_bicluster_0072 | 0.37 | Thalassiosira pseudonana |
Displaying 1 - 46 of 46
" class="views-fluidgrid-wrapper clear-block">
10010 hypothetical protein
10023 hypothetical protein
10509 hypothetical protein
10713 hypothetical protein
10754 hypothetical protein
11052 hypothetical protein
11403 hypothetical protein
11407 hypothetical protein
11408 hypothetical protein
11536 DDE_Tnp_1_7 superfamily
11541 hypothetical protein
11649 hypothetical protein
11681 hypothetical protein
12124 hypothetical protein
12125 hypothetical protein
1324 DDE_Tnp_1_7 superfamily
1353 hypothetical protein
2365 hypothetical protein
24978 hypothetical protein
25550 hypothetical protein
262191 RNase_HI_RT_Ty1
263849 FG-GAP_2
263938 FG-GAP_2
264479 FG-GAP_2
264557 FG-GAP_2
264660 FG-GAP_2
264954 FG-GAP_2
3030 hypothetical protein
5844 hypothetical protein
6604 hypothetical protein
6879 hypothetical protein
7300 hypothetical protein
7603 hypothetical protein
7608 hypothetical protein
7638 hypothetical protein
8289 RVT_2 superfamily
8290 hypothetical protein
8608 hypothetical protein
867 hypothetical protein
8795 hypothetical protein
8874 hypothetical protein
9153 hypothetical protein
9642 hypothetical protein
| GO ID | Go Term | p-value | q-value | Cluster |
|---|---|---|---|---|
| GO:0007160 | cell-matrix adhesion | 0.00000000000309 | 0.00000000857 | Thaps_bicluster_0072 |
| GO:0007155 | cell adhesion | 0.0000000000972 | 0.000000269 | Thaps_bicluster_0072 |
| GO:0006310 | DNA recombination | 0.0536808 | 1 | Thaps_bicluster_0072 |
| GO:0008152 | metabolism | 0.708368 | 1 | Thaps_bicluster_0072 |

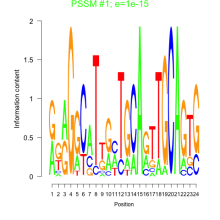
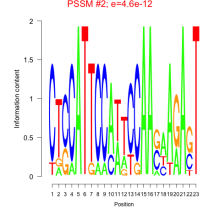
Comments