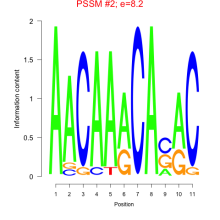
Thaps_bicluster_0073 Residual: 0.47
Thalassiosira pseudonana
| Title | Residual | Model version | Organism |
|---|---|---|---|
| Thaps_bicluster_0073 | 0.47 | Thalassiosira pseudonana |
Displaying 1 - 41 of 41
" class="views-fluidgrid-wrapper clear-block">
10482 hypothetical protein
10749 M36
10929 NYD-SP28
11534 hypothetical protein
1173 (Tp_HSF_2.7f) regulator [Rayko]
1303 WD40 superfamily
1588 hypothetical protein
16501 PKc_like superfamily
1740 PDZ superfamily
1798 hypothetical protein
1900 hypothetical protein
20674 WD40 superfamily
21133 COG3899
21203 hypothetical protein
21655 hypothetical protein
2320 hypothetical protein
23598 COG3899
24029 hypothetical protein
24267 FPL superfamily
24515 hypothetical protein
261023 hypothetical protein
261181 Gly_transf_sug superfamily
262880 IlvB
262923 Peptidase_M8 superfamily
263642 (CDA1) CE4_PuuE_SpCDA1
264262 COG3899
264726 REC
33497 MEMO_like
35323 RRM_SR140
35373 AarF
3590 Ion_trans_2
3947 hypothetical protein
5763 ERCC4
6560 COG3899
6726 hypothetical protein
7553 hypothetical protein
7582 CHD
7935 Sulfotransfer_2 superfamily
7989 globin_like superfamily
8510 hypothetical protein
9653 M36
| GO ID | Go Term | p-value | q-value | Cluster |
|---|---|---|---|---|
| GO:0015671 | oxygen transport | 0.00413479 | 1 | Thaps_bicluster_0073 |
| GO:0006468 | protein amino acid phosphorylation | 0.0552342 | 1 | Thaps_bicluster_0073 |
| GO:0009190 | cyclic nucleotide biosynthesis | 0.0873045 | 1 | Thaps_bicluster_0073 |
| GO:0006813 | potassium ion transport | 0.0873045 | 1 | Thaps_bicluster_0073 |
| GO:0000160 | two-component signal transduction system (phosphorelay) | 0.106111 | 1 | Thaps_bicluster_0073 |
| GO:0006508 | proteolysis and peptidolysis | 0.112621 | 1 | Thaps_bicluster_0073 |
| GO:0007155 | cell adhesion | 0.117217 | 1 | Thaps_bicluster_0073 |
| GO:0007242 | intracellular signaling cascade | 0.160345 | 1 | Thaps_bicluster_0073 |
| GO:0006396 | RNA processing | 0.214744 | 1 | Thaps_bicluster_0073 |
| GO:0008152 | metabolism | 0.234081 | 1 | Thaps_bicluster_0073 |


Comments