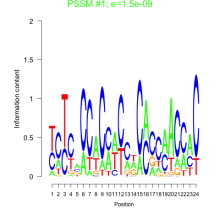
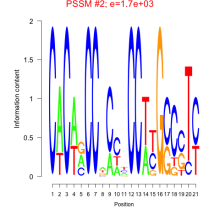
Thaps_bicluster_0074 Residual: 0.37
Thalassiosira pseudonana
| Title | Residual | Model version | Organism |
|---|---|---|---|
| Thaps_bicluster_0074 | 0.37 | Thalassiosira pseudonana |
Displaying 1 - 55 of 55
" class="views-fluidgrid-wrapper clear-block">
10035 hypothetical protein
10082 hypothetical protein
10099 HMGB-UBF_HMG-box
10468 TMP_TenI
10961 hypothetical protein
11088 P4Hc
11252 hypothetical protein
11253 DDE_Tnp_4
11330 hypothetical protein
11515 (Tp_HSF_2.5f) HSF_DNA-bind superfamily
11647 hypothetical protein
11650 hypothetical protein
11758 hypothetical protein
11896 hypothetical protein
12126 hypothetical protein
1947 hypothetical protein
2001 hypothetical protein
20330 zf-Tim10_DDP
2042 hypothetical protein
2100 hypothetical protein
21363 MRP-S28 superfamily
22026 hypothetical protein
24954 hypothetical protein
263627 FG-GAP_2
263745 PDEase_I
33377 (cycB) CYCLIN
38726 hypothetical protein
38886 zf-Tim10_DDP
40767 DUF3523
4431 Rhamno_transf superfamily
4483 hypothetical protein
4813 hypothetical protein
6645 hypothetical protein
6722 hypothetical protein
7244 hypothetical protein
7343 (Tp_HSF_2.3b) HSF_DNA-bind superfamily
7554 hypothetical protein
7768 hypothetical protein
8075 DENN
8233 hypothetical protein
8236 hypothetical protein
8517 hypothetical protein
879 hypothetical protein
8895 hypothetical protein
8898 hypothetical protein
8900 hypothetical protein
9032 hypothetical protein
9161 DDE_Tnp_1_7 superfamily
9279 hypothetical protein
9404 hypothetical protein
9867 LbetaH superfamily
| GO ID | Go Term | p-value | q-value | Cluster |
|---|---|---|---|---|
| GO:0045039 | mitochondrial inner membrane protein import | 0.0000695 | 0.191 | Thaps_bicluster_0074 |
| GO:0006626 | protein-mitochondrial targeting | 0.0000695 | 0.191 | Thaps_bicluster_0074 |
| GO:0019538 | protein metabolism | 0.0109894 | 1 | Thaps_bicluster_0074 |
| GO:0009228 | thiamin biosynthesis | 0.0139887 | 1 | Thaps_bicluster_0074 |
| GO:0006355 | regulation of transcription, DNA-dependent | 0.0195945 | 1 | Thaps_bicluster_0074 |
| GO:0006508 | proteolysis | 0.0481589 | 1 | Thaps_bicluster_0074 |
| GO:0009401 | phosphoenolpyruvate-dependent sugar phosphotransferase system | 0.0615124 | 1 | Thaps_bicluster_0074 |
| GO:0007160 | cell-matrix adhesion | 0.0615124 | 1 | Thaps_bicluster_0074 |
| GO:0007155 | cell adhesion | 0.100523 | 1 | Thaps_bicluster_0074 |
| GO:0007165 | signal transduction | 0.13186 | 1 | Thaps_bicluster_0074 |

Comments