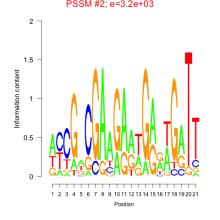
Thaps_bicluster_0081 Residual: 0.42
Thalassiosira pseudonana
| Title | Residual | Model version | Organism |
|---|---|---|---|
| Thaps_bicluster_0081 | 0.42 | Thalassiosira pseudonana |
Displaying 1 - 22 of 22
" class="views-fluidgrid-wrapper clear-block">
10871 hypothetical protein
11462 hypothetical protein
1665 hypothetical protein
22890 ERO1 superfamily
23043 hypothetical protein
24082 Methyltransf_31
25281 NADB_Rossmann superfamily
25707 P4Hc
262069 KefB
263661 Mito_carr
26530 CzcD
268124 COG0429
270206 (Tpt8) hypothetical protein
30928 PcbC
3105 Glyco_hydrolase_16 superfamily
31907 ipk
37444 PLN02244
37516 Mpv17_PMP22
6797 hypothetical protein
8042 hypothetical protein
923 hypothetical protein
9401 hypothetical protein
| GO ID | Go Term | p-value | q-value | Cluster |
|---|---|---|---|---|
| GO:0016114 | terpenoid biosynthesis | 0.00577795 | 1 | Thaps_bicluster_0081 |
| GO:0017000 | antibiotic biosynthesis | 0.00721795 | 1 | Thaps_bicluster_0081 |
| GO:0016310 | phosphorylation | 0.0186738 | 1 | Thaps_bicluster_0081 |
| GO:0006812 | cation transport | 0.06063 | 1 | Thaps_bicluster_0081 |
| GO:0019538 | protein metabolism | 0.0647398 | 1 | Thaps_bicluster_0081 |
| GO:0006810 | transport | 0.237879 | 1 | Thaps_bicluster_0081 |
| GO:0006118 | electron transport | 0.327122 | 1 | Thaps_bicluster_0081 |

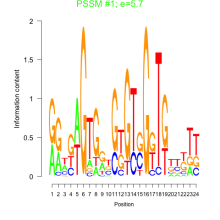
Comments