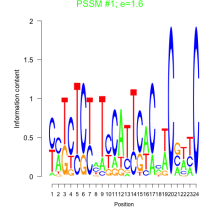
Thaps_bicluster_0083 Residual: 0.43
Thalassiosira pseudonana
| Title | Residual | Model version | Organism |
|---|---|---|---|
| Thaps_bicluster_0083 | 0.43 | Thalassiosira pseudonana |
Displaying 1 - 32 of 32
" class="views-fluidgrid-wrapper clear-block">
10693 hypothetical protein
11314 hypothetical protein
16828 S_TKc
17845 RRM_CNOT4
18076 ADK
21104 Glyco_hydrolase_16 superfamily
21612 PRK07206
21809 hypothetical protein
2220 (Tp_HSF_2.4e) regulator [Rayko]
22429 hypothetical protein
24261 hypothetical protein
25037 hypothetical protein
25462 Ferritin_like superfamily
25755 hypothetical protein
25868 Methyltransf_16 superfamily
26051 (GLNA) PLN02284
262025 GH18_chitinase-like superfamily
262083 Uup
262152 hypothetical protein
263459 MATE_like superfamily
26560 PLN03026
28441 eIF-3c_N superfamily
32477 (ODC1) PLPDE_III_ODC
33968 (CEN2) PTZ00183
34348 SAM_decarbox superfamily
3563 (Tp_CCCH2) WD40 superfamily
36614 PLN02536
41122 MPN_eIF3h
4740 (Tp_HSF_2.2c) HSF_DNA-bind superfamily
5885 Methyltrans_SAM
5899 hypothetical protein
ThpsCt009 (trnS(gcu))
| GO ID | Go Term | p-value | q-value | Cluster |
|---|---|---|---|---|
| GO:0006542 | glutamine biosynthesis | 0.00434153 | 1 | Thaps_bicluster_0083 |
| GO:0006596 | polyamine biosynthesis | 0.00434153 | 1 | Thaps_bicluster_0083 |
| GO:0008295 | spermidine biosynthesis | 0.00434153 | 1 | Thaps_bicluster_0083 |
| GO:0006597 | spermine biosynthesis | 0.00434153 | 1 | Thaps_bicluster_0083 |
| GO:0009089 | lysine biosynthesis via diaminopimelate | 0.0129708 | 1 | Thaps_bicluster_0083 |
| GO:0001539 | ciliary or flagellar motility | 0.0172587 | 1 | Thaps_bicluster_0083 |
| GO:0000105 | histidine biosynthesis | 0.0342335 | 1 | Thaps_bicluster_0083 |
| GO:0006807 | nitrogen compound metabolism | 0.0467809 | 1 | Thaps_bicluster_0083 |
| GO:0006139 | nucleobase, nucleoside, nucleotide and nucleic acid metabolism | 0.0632695 | 1 | Thaps_bicluster_0083 |
| GO:0006413 | translational initiation | 0.0993813 | 1 | Thaps_bicluster_0083 |


Comments