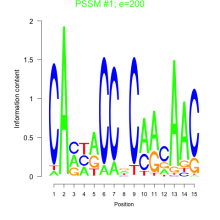
Thaps_bicluster_0087 Residual: 0.32
Thalassiosira pseudonana
| Title | Residual | Model version | Organism |
|---|---|---|---|
| Thaps_bicluster_0087 | 0.32 | Thalassiosira pseudonana |
Displaying 1 - 38 of 38
" class="views-fluidgrid-wrapper clear-block">
11063 hypothetical protein
11103 hypothetical protein
11254 hypothetical protein
11255 hypothetical protein
11488 hypothetical protein
11537 hypothetical protein
11720 hypothetical protein
12147 hypothetical protein
21549 hypothetical protein
21869 hypothetical protein
22961 NPD_like
23371 PRK09358
24018 hypothetical protein
24195 HA
260805 SRP54_euk
2612 hypothetical protein
263580 (Tp_CSF1) regulator [Rayko]
263793 hypothetical protein
263840 hypothetical protein
264582 lysozyme_like superfamily
268213 hypothetical protein
269474 (Tp_HSF_1.2c) HSF_DNA-bind
2755 hypothetical protein
2886 hypothetical protein
30683 proteasome_alpha_type_2
37485 Abhydrolase_5
40627 MPP_PP2A_PP4_PP6
4422 hypothetical protein
5897 hypothetical protein
6571 hypothetical protein
6705 UPF0029 superfamily
7699 COG2391
7743 hypothetical protein
8183 hypothetical protein
8835 hypothetical protein
8962 2a58
9298 hypothetical protein
| GO ID | Go Term | p-value | q-value | Cluster |
|---|---|---|---|---|
| GO:0009168 | purine ribonucleoside monophosphate biosynthesis | 0.00680831 | 1 | Thaps_bicluster_0087 |
| GO:0006817 | phosphate transport | 0.0113237 | 1 | Thaps_bicluster_0087 |
| GO:0006614 | SRP-dependent cotranslational protein-membrane targeting | 0.0135745 | 1 | Thaps_bicluster_0087 |
| GO:0006605 | protein targeting | 0.0158205 | 1 | Thaps_bicluster_0087 |
| GO:0006032 | chitin catabolism | 0.0180619 | 1 | Thaps_bicluster_0087 |
| GO:0016998 | cell wall catabolism | 0.0202987 | 1 | Thaps_bicluster_0087 |
| GO:0009613 | response to pest, pathogen or parasite | 0.0202987 | 1 | Thaps_bicluster_0087 |
| GO:0006511 | ubiquitin-dependent protein catabolism | 0.110164 | 1 | Thaps_bicluster_0087 |
| GO:0006355 | regulation of transcription, DNA-dependent | 0.151344 | 1 | Thaps_bicluster_0087 |
| GO:0006118 | electron transport | 0.463588 | 1 | Thaps_bicluster_0087 |

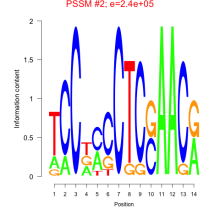
Comments