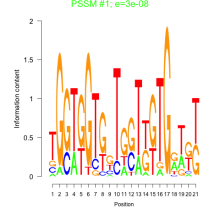
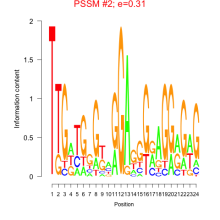
Thaps_bicluster_0091 Residual: 0.31
Thalassiosira pseudonana
| Title | Residual | Model version | Organism |
|---|---|---|---|
| Thaps_bicluster_0091 | 0.31 | Thalassiosira pseudonana |
Displaying 1 - 20 of 20
" class="views-fluidgrid-wrapper clear-block">
1122 hypothetical protein
1357 FKBP_C
19301 sigpep_I_bact
20653 PseudoU_synth superfamily
20734 hypothetical protein
21176 FMO-like
22417 Sun
22847 hypothetical protein
23246 hypothetical protein
25365 hypothetical protein
262503 Nudix_Hydrolase superfamily
269034 hypothetical protein
269798 hypothetical protein
31627 RluA
31819 DUF2419 superfamily
3292 COG2319
35297 AdoMet_MTases superfamily
36917 TGT superfamily
5241 RNase_P_p30 superfamily
8261 DUF819 superfamily
| GO ID | Go Term | p-value | q-value | Cluster |
|---|---|---|---|---|
| GO:0008616 | queuosine biosynthesis | 0.00660132 | 1 | Thaps_bicluster_0091 |
| GO:0006400 | tRNA modification | 0.00660132 | 1 | Thaps_bicluster_0091 |
| GO:0006396 | RNA processing | 0.0920603 | 1 | Thaps_bicluster_0091 |
| GO:0006457 | protein folding | 0.195964 | 1 | Thaps_bicluster_0091 |
| GO:0006118 | electron transport | 0.364179 | 1 | Thaps_bicluster_0091 |
| GO:0006508 | proteolysis and peptidolysis | 0.38827 | 1 | Thaps_bicluster_0091 |
| GO:0006355 | regulation of transcription, DNA-dependent | 0.40843 | 1 | Thaps_bicluster_0091 |
| GO:0008152 | metabolism | 0.505245 | 1 | Thaps_bicluster_0091 |

Comments