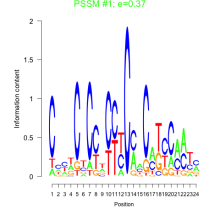
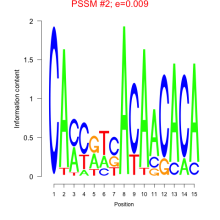
Thaps_bicluster_0097 Residual: 0.39
Thalassiosira pseudonana
| Title | Residual | Model version | Organism |
|---|---|---|---|
| Thaps_bicluster_0097 | 0.39 | Thalassiosira pseudonana |
Displaying 1 - 48 of 48
" class="views-fluidgrid-wrapper clear-block">
10178 hypothetical protein
10207 hypothetical protein
11114 UPF0004
1134 DSPc superfamily
1412 TraB superfamily
1563 hypothetical protein
1629 hypothetical protein
1882 Gn_AT_II superfamily
19081 hypothetical protein
20431 zf-CHC2 superfamily
20504 cyclophilin superfamily
21750 Octopine_DH superfamily
2244 hypothetical protein
22801 hypothetical protein
23622 RhaT
24218 hypothetical protein
24451 TFIIS_I superfamily
24615 hypothetical protein
261325 ARM
261876 (ZFP9) hypothetical protein
261998 (FRU3) frustulin protein
262512 hypothetical protein
262593 (MTH1_2) Nudix_Hydrolase superfamily
262844 heat shock protein
267977 MATE_like superfamily
268179 DTW superfamily
268845 COG3899
269985 ELFV_dehydrog_N
2959 hypothetical protein
31928 ATM1
32154 (infC) IF3_C superfamily
32486 HA
33600 PTZ00184
33726 STKc_CDK9_like
3674 hypothetical protein
38466 SpoU_methylase superfamily
38752 hypothetical protein
40473 AMMECR1
5488 CAP_ED
5779 hypothetical protein
7419 hypothetical protein
7804 MATE_like superfamily
7870 hypothetical protein
8161 hypothetical protein
8924 hypothetical protein
9103 hypothetical protein
9125 hypothetical protein
| GO ID | Go Term | p-value | q-value | Cluster |
|---|---|---|---|---|
| GO:0006754 | ATP biosynthetic process | 0.00289436 | 1 | Thaps_bicluster_0097 |
| GO:0015986 | ATP synthesis coupled proton transport | 0.125374 | 1 | Thaps_bicluster_0097 |
| GO:0005975 | carbohydrate metabolic process | 0.0143942 | 1 | Thaps_bicluster_0097 |
| GO:0006260 | DNA replication | 0.0967972 | 1 | Thaps_bicluster_0097 |
| GO:0006811 | ion transport | 0.0509236 | 1 | Thaps_bicluster_0097 |
| GO:0008152 | metabolism | 0.708368 | 1 | Thaps_bicluster_0097 |
| GO:0006470 | protein amino acid dephosphorylation | 0.0727741 | 1 | Thaps_bicluster_0097 |
| GO:0006468 | protein amino acid phosphorylation | 0.468128 | 1 | Thaps_bicluster_0097 |
| GO:0006457 | protein folding | 0.0528208 | 1 | Thaps_bicluster_0097 |
| GO:0006355 | regulation of transcription, DNA-dependent | 0.6012 | 1 | Thaps_bicluster_0097 |

Comments