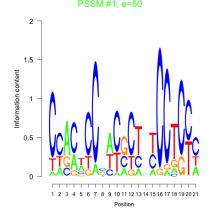
Thaps_bicluster_0113 Residual: 0.35
Thalassiosira pseudonana
| Title | Residual | Model version | Organism |
|---|---|---|---|
| Thaps_bicluster_0113 | 0.35 | Thalassiosira pseudonana |
Displaying 1 - 27 of 27
" class="views-fluidgrid-wrapper clear-block">
11718 hypothetical protein
11762 hypothetical protein
11902 RPA_2b-aaRSs_OBF_like superfamily
12172 hypothetical protein
1765 hypothetical protein
18434 PIPKc superfamily
22112 Vps54 superfamily
2224 hypothetical protein
22916 DUF1162 superfamily
2314 hypothetical protein
23614 hypothetical protein
23952 hypothetical protein
2502 hypothetical protein
261754 SUL1
264058 phosphatase 2A (PP2A) regulatory subunit B-like protein-like protein
33092 RINGv superfamily
3611 MIT_CorA-like superfamily
3751 hypothetical protein
37942 hypothetical protein
41637 Patatin_and_cPLA2 superfamily
5010 (Tp_HSF_3.6b) HSF_DNA-bind superfamily
6784 hypothetical protein
7633 FPL superfamily
8576 Ion_trans_2
9431 DUF399 superfamily
9491 hypothetical protein
9693 hypothetical protein
| GO ID | Go Term | p-value | q-value | Cluster |
|---|---|---|---|---|
| GO:0006813 | potassium ion transport | 0.0135857 | 1 | Thaps_bicluster_0113 |
| GO:0016567 | protein ubiquitination | 0.0482112 | 1 | Thaps_bicluster_0113 |
| GO:0006355 | regulation of transcription, DNA-dependent | 0.178613 | 1 | Thaps_bicluster_0113 |

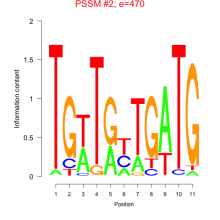
Comments