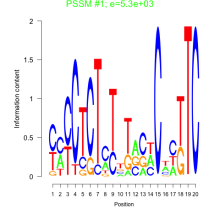
Thaps_bicluster_0134 Residual: 0.26
Thalassiosira pseudonana
| Title | Residual | Model version | Organism |
|---|---|---|---|
| Thaps_bicluster_0134 | 0.26 | Thalassiosira pseudonana |
Displaying 1 - 15 of 15
" class="views-fluidgrid-wrapper clear-block">
10002 Solute_trans_a superfamily
11492 hypothetical protein
21094 hypothetical protein
22301 (PMM1) PLN02423
22325 hypothetical protein
25772 (ACT1) ACTIN
25898 hypothetical protein
25917 hypothetical protein
261826 SEC14
31569 (TUB3) PLN00220
32661 ABC_ATPase superfamily
4170 hypothetical protein
4803 hypothetical protein
5431 hypothetical protein
5867 CDC6
| GO ID | Go Term | p-value | q-value | Cluster |
|---|---|---|---|---|
| GO:0019307 | mannose biosynthesis | 0.0010337 | 1 | Thaps_bicluster_0134 |
| GO:0008152 | metabolism | 0.355748 | 1 | Thaps_bicluster_0134 |
| GO:0007018 | microtubule-based movement | 0.0436835 | 1 | Thaps_bicluster_0134 |
| GO:0006298 | mismatch repair | 0.0164369 | 1 | Thaps_bicluster_0134 |
| GO:0051258 | protein polymerization | 0.00824568 | 1 | Thaps_bicluster_0134 |


Comments