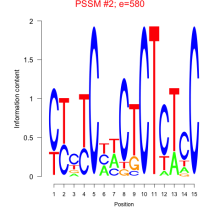
Thaps_bicluster_0135 Residual: 0.41
Thalassiosira pseudonana
| Title | Residual | Model version | Organism |
|---|---|---|---|
| Thaps_bicluster_0135 | 0.41 | Thalassiosira pseudonana |
Displaying 1 - 38 of 38
" class="views-fluidgrid-wrapper clear-block">
11145 hypothetical protein
11367 GH_J
11624 Fip1
18820 (Tp_Myb4R1a) SANT
20671 hypothetical protein
21544 hypothetical protein
21776 hypothetical protein
22640 hypothetical protein
22765 hypothetical protein
24655 hypothetical protein
24722 hypothetical protein
25064 hypothetical protein
25217 hypothetical protein
25290 hypothetical protein
25426 hypothetical protein
261067 alkPPc superfamily
261613 SPRY_RanBP_like
264335 PIP5K
264395 PI3Kc_like superfamily
268194 (Tp_Myb4R1b) SANT
268895 (SIT1) Silic_transp superfamily
29771 PIKKc_TOR
35963 CE4_SF superfamily
36322 TrmA
3706 hypothetical protein
3781 hypothetical protein
40958 (TPI1) TIM
41392 (SIT2) Silic_transp superfamily
4355 hypothetical protein
4755 GST_C_family superfamily
4825 hypothetical protein
4958 hypothetical protein
5708 hypothetical protein
6880 hypothetical protein
8372 hypothetical protein
9222 COG3899
9479 ATG22
9619 hypothetical protein
| GO ID | Go Term | p-value | q-value | Cluster |
|---|---|---|---|---|
| GO:0051336 | regulation of hydrolase activity | 0.0129601 | 1 | Thaps_bicluster_0135 |
| GO:0030163 | protein catabolism | 0.0239497 | 1 | Thaps_bicluster_0135 |
| GO:0008152 | metabolism | 0.0338556 | 1 | Thaps_bicluster_0135 |
| GO:0006512 | ubiquitin cycle | 0.065076 | 1 | Thaps_bicluster_0135 |
| GO:0006396 | RNA processing | 0.102965 | 1 | Thaps_bicluster_0135 |
| GO:0006464 | protein modification | 0.104655 | 1 | Thaps_bicluster_0135 |
| GO:0006118 | electron transport | 0.399203 | 1 | Thaps_bicluster_0135 |


Comments