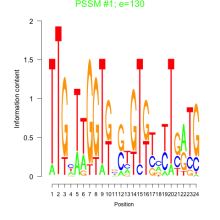
Thaps_bicluster_0137 Residual: 0.33
Thalassiosira pseudonana
| Title | Residual | Model version | Organism |
|---|---|---|---|
| Thaps_bicluster_0137 | 0.33 | Thalassiosira pseudonana |
Displaying 1 - 22 of 22
" class="views-fluidgrid-wrapper clear-block">
20014 TBCA
22075 Hydrolase_like superfamily
22464 (FUM1) PLN00133
22792 Rab1_Ypt1
2409 MPN_NPL4
262994 COG0637
263375 RpoD_Cterm
264773 ABCG_PDR_domain2
269540 (CHC) Clathrin
30971 MIR
3500 hypothetical protein
42475 PLN00128
4704 LPLAT_AAK14816-like
4783 hypothetical protein
5586 Prenylcys_lyase superfamily
5599 HVSL superfamily
5701 hypothetical protein
6552 hypothetical protein
8353 hypothetical protein
889 hypothetical protein
904 hypothetical protein
9848 Thioredoxin_like superfamily
| GO ID | Go Term | p-value | q-value | Cluster |
|---|---|---|---|---|
| GO:0007022 | chaperonin-mediated tubulin folding | 0.00289436 | 1 | Thaps_bicluster_0137 |
| GO:0006091 | generation of precursor metabolites and energy | 0.00289436 | 1 | Thaps_bicluster_0137 |
| GO:0006099 | tricarboxylic acid cycle | 0.0285957 | 1 | Thaps_bicluster_0137 |
| GO:0006352 | transcription initiation | 0.0370252 | 1 | Thaps_bicluster_0137 |
| GO:0000160 | two-component signal transduction system (phosphorelay) | 0.0754724 | 1 | Thaps_bicluster_0137 |
| GO:0007264 | small GTPase mediated signal transduction | 0.0941568 | 1 | Thaps_bicluster_0137 |
| GO:0007018 | microtubule-based movement | 0.117664 | 1 | Thaps_bicluster_0137 |
| GO:0015031 | protein transport | 0.117664 | 1 | Thaps_bicluster_0137 |
| GO:0006886 | intracellular protein transport | 0.145627 | 1 | Thaps_bicluster_0137 |
| GO:0006355 | regulation of transcription, DNA-dependent | 0.221736 | 1 | Thaps_bicluster_0137 |

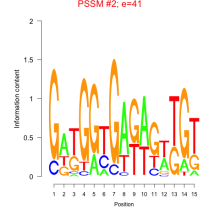
Comments