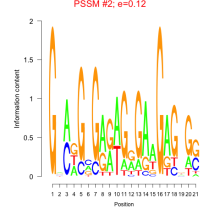
Thaps_bicluster_0153 Residual: 0.31
Thalassiosira pseudonana
| Title | Residual | Model version | Organism |
|---|---|---|---|
| Thaps_bicluster_0153 | 0.31 | Thalassiosira pseudonana |
Displaying 1 - 23 of 23
" class="views-fluidgrid-wrapper clear-block">
20974 hypothetical protein
21333 Reprolysin_3
21438 hypothetical protein
22215 hypothetical protein
22708 hypothetical protein
23314 hypothetical protein
23419 M6dom_TIGR03296
23521 hypothetical protein
23933 hypothetical protein
23947 hypothetical protein
23949 hypothetical protein
262247 (CHT2) Glyco_18
263096 lysozyme_like superfamily
269264 hypothetical protein
2693 hypothetical protein
32693 hypothetical protein
34418 Phytochelatin
4193 hypothetical protein
6273 LRR_RI superfamily
6424 hypothetical protein
6750 hypothetical protein
7333 hypothetical protein
9406 hypothetical protein
| GO ID | Go Term | p-value | q-value | Cluster |
|---|---|---|---|---|
| GO:0006030 | chitin metabolism | 0.0000114 | 0.0314097 | Thaps_bicluster_0153 |
| GO:0006032 | chitin catabolism | 0.0180619 | 1 | Thaps_bicluster_0153 |
| GO:0016998 | cell wall catabolism | 0.0202987 | 1 | Thaps_bicluster_0153 |
| GO:0009613 | response to pest, pathogen or parasite | 0.0202987 | 1 | Thaps_bicluster_0153 |
| GO:0006508 | proteolysis and peptidolysis | 0.0241111 | 1 | Thaps_bicluster_0153 |
| GO:0005975 | carbohydrate metabolism | 0.158085 | 1 | Thaps_bicluster_0153 |
| GO:0008152 | metabolism | 0.620117 | 1 | Thaps_bicluster_0153 |


Comments