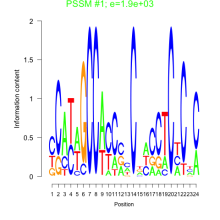
Thaps_bicluster_0154 Residual: 0.36
Thalassiosira pseudonana
| Title | Residual | Model version | Organism |
|---|---|---|---|
| Thaps_bicluster_0154 | 0.36 | Thalassiosira pseudonana |
Displaying 1 - 30 of 30
" class="views-fluidgrid-wrapper clear-block">
11466 hypothetical protein
13089 Uup
21390 PS_II_psbQ_bact superfamily
22042 DUF676
22807 Methyltransf_4 superfamily
23215 Ubox
23368 hypothetical protein
23446 hypothetical protein
23662 Tryp_SPc
23720 hypothetical protein
24198 hypothetical protein
24537 hypothetical protein
24677 hypothetical protein
25069 hypothetical protein
25091 hypothetical protein
25092 hypothetical protein
25107 U-box superfamily
25206 hypothetical protein
25207 hypothetical protein
263451 Tryp_SPc
263452 Tryp_SPc
264805 hypothetical protein
268204 hypothetical protein
31472 S_TKc
32738 S_TKc
35259 hypothetical protein
36568 COG2119
39824 (HSP104) chaperone_ClpB
42804 Tryp_SPc
5770 hypothetical protein
| GO ID | Go Term | p-value | q-value | Cluster |
|---|---|---|---|---|
| GO:0006508 | proteolysis and peptidolysis | 0.00416092 | 1 | Thaps_bicluster_0154 |
| GO:0016567 | protein ubiquitination | 0.015635 | 1 | Thaps_bicluster_0154 |
| GO:0006298 | mismatch repair | 0.0390234 | 1 | Thaps_bicluster_0154 |
| GO:0006468 | protein amino acid phosphorylation | 0.0953042 | 1 | Thaps_bicluster_0154 |
| GO:0019538 | protein metabolism | 0.108453 | 1 | Thaps_bicluster_0154 |
| GO:0006457 | protein folding | 0.279136 | 1 | Thaps_bicluster_0154 |
| GO:0006355 | regulation of transcription, DNA-dependent | 0.545155 | 1 | Thaps_bicluster_0154 |


Comments