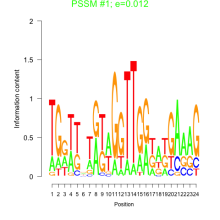
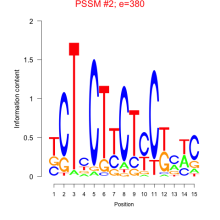
Thaps_bicluster_0158 Residual: 0.37
Thalassiosira pseudonana
| Title | Residual | Model version | Organism |
|---|---|---|---|
| Thaps_bicluster_0158 | 0.37 | Thalassiosira pseudonana |
Displaying 1 - 28 of 28
" class="views-fluidgrid-wrapper clear-block">
10814 hypothetical protein
10816 hypothetical protein
11038 hypothetical protein
11039 TLD
11057 hypothetical protein
12149 hypothetical protein
12960 Vac14_Fig4_bd
20644 hypothetical protein
21861 hypothetical protein
22070 SRPBCC superfamily
23881 hypothetical protein
24101 hypothetical protein
24235 Peptidase_C19 superfamily
24870 hypothetical protein
25181 hypothetical protein
25429 hypothetical protein
25882 hypothetical protein
268835 (ECI1) ECH
269233 (Tp_HSF_3.4d) HSF_DNA-bind
3625 FNR_like superfamily
38190 VTC1
4157 hypothetical protein
6636 Glyco_hydro_18
6798 PRK06476
8980 hypothetical protein
9657 hypothetical protein
9661 hypothetical protein
9959 hypothetical protein
| GO ID | Go Term | p-value | q-value | Cluster |
|---|---|---|---|---|
| GO:0005975 | carbohydrate metabolism | 0.131302 | 1 | Thaps_bicluster_0158 |
| GO:0006030 | chitin metabolism | 0.0384333 | 1 | Thaps_bicluster_0158 |
| GO:0006118 | electron transport | 0.399203 | 1 | Thaps_bicluster_0158 |
| GO:0008152 | metabolism | 0.171652 | 1 | Thaps_bicluster_0158 |
| GO:0006561 | proline biosynthesis | 0.00742418 | 1 | Thaps_bicluster_0158 |
| GO:0006355 | regulation of transcription, DNA-dependent | 0.446038 | 1 | Thaps_bicluster_0158 |
| GO:0006512 | ubiquitin cycle | 0.065076 | 1 | Thaps_bicluster_0158 |
| GO:0006511 | ubiquitin-dependent protein catabolism | 0.0910605 | 1 | Thaps_bicluster_0158 |

Comments