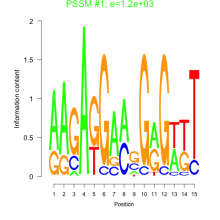
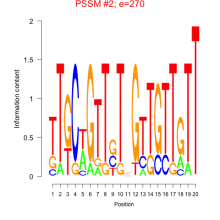
Thaps_bicluster_0170 Residual: 0.34
Thalassiosira pseudonana
| Title | Residual | Model version | Organism |
|---|---|---|---|
| Thaps_bicluster_0170 | 0.34 | Thalassiosira pseudonana |
Displaying 1 - 26 of 26
" class="views-fluidgrid-wrapper clear-block">
10937 hypothetical protein
11360 hypothetical protein
11984 hypothetical protein
20970 SpoVK
21085 hypothetical protein
21344 TPP_enzyme_PYR superfamily
21515 hypothetical protein
21558 hypothetical protein
21656 hypothetical protein
23511 hypothetical protein
23576 HA
23671 hypothetical protein
24428 hypothetical protein
24596 PKc
24900 hypothetical protein
25637 hypothetical protein
25783 hypothetical protein
263048 hypothetical protein
269115 hypothetical protein
31001 Y_phosphoryl
31984 (SAT2) cysE
37231 HECTc
37803 DUF208 superfamily
39225 Snf7
7550 hypothetical protein
7689 hypothetical protein
| GO ID | Go Term | p-value | q-value | Cluster |
|---|---|---|---|---|
| GO:0006512 | ubiquitin cycle | 0.0509804 | 1 | Thaps_bicluster_0170 |
| GO:0006206 | pyrimidine base metabolism | 0.00144718 | 1 | Thaps_bicluster_0170 |
| GO:0006464 | protein modification | 0.0823711 | 1 | Thaps_bicluster_0170 |
| GO:0006468 | protein amino acid phosphorylation | 0.270534 | 1 | Thaps_bicluster_0170 |
| GO:0008152 | metabolism | 0.0160137 | 1 | Thaps_bicluster_0170 |

Comments