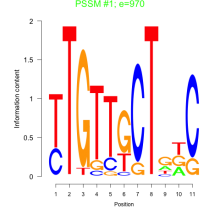
Thaps_bicluster_0179 Residual: 0.28
Thalassiosira pseudonana
| Title | Residual | Model version | Organism |
|---|---|---|---|
| Thaps_bicluster_0179 | 0.28 | Thalassiosira pseudonana |
Displaying 1 - 15 of 15
" class="views-fluidgrid-wrapper clear-block">
21709 hypothetical protein
22356 PseudoU_synth_ScPUS7
22965 SrmB
23812 TrmA
24015 Urb2 superfamily
24038 hypothetical protein
260748 AdoMet_MTases superfamily
264188 COG1026
268412 RluA
269080 PLN02327
269917 TrmA
32668 CRCB
32882 PRK15440
34596 WD40
5794 hypothetical protein
| GO ID | Go Term | p-value | q-value | Cluster |
|---|---|---|---|---|
| GO:0008033 | tRNA processing | 0.000271696 | 0.742002 | Thaps_bicluster_0179 |
| GO:0006221 | pyrimidine nucleotide biosynthesis | 0.00433615 | 1 | Thaps_bicluster_0179 |
| GO:0006396 | RNA processing | 0.081025 | 1 | Thaps_bicluster_0179 |
| GO:0008152 | metabolism | 0.111784 | 1 | Thaps_bicluster_0179 |
| GO:0006508 | proteolysis and peptidolysis | 0.349482 | 1 | Thaps_bicluster_0179 |

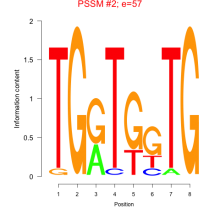
Comments