Thaps_bicluster_0186 Residual: 0.38
Thalassiosira pseudonana
| Title | Residual | Model version | Organism |
|---|---|---|---|
| Thaps_bicluster_0186 | 0.38 | Thalassiosira pseudonana |
Displaying 1 - 22 of 22
" class="views-fluidgrid-wrapper clear-block">
10271 hypothetical protein
12943 NARP1
15688 MFS
22750 hypothetical protein
23106 hypothetical protein
23472 hypothetical protein
24140 hypothetical protein
24632 PDCD2_C superfamily
2609 WD40 superfamily
261250 Phytochelatin
262343 LacZ
262400 ANK
269376 SrmB
3017 SNARE_assoc
34710 SNF2_N
36345 (Tp_sigma70.3b) Sig70-cyanoRpoD
36501 UBCc superfamily
38475 HA2
38757 trmE
5053 hypothetical protein
6255 hypothetical protein
8259 hypothetical protein
| GO ID | Go Term | p-value | q-value | Cluster |
|---|---|---|---|---|
| GO:0006352 | transcription initiation | 0.0265779 | 1 | Thaps_bicluster_0186 |
| GO:0006310 | DNA recombination | 0.0386288 | 1 | Thaps_bicluster_0186 |
| GO:0006260 | DNA replication | 0.0701108 | 1 | Thaps_bicluster_0186 |
| GO:0006512 | ubiquitin cycle | 0.0720473 | 1 | Thaps_bicluster_0186 |
| GO:0019538 | protein metabolism | 0.0912132 | 1 | Thaps_bicluster_0186 |
| GO:0006281 | DNA repair | 0.0912132 | 1 | Thaps_bicluster_0186 |
| GO:0006464 | protein modification | 0.115596 | 1 | Thaps_bicluster_0186 |
| GO:0006810 | transport | 0.321719 | 1 | Thaps_bicluster_0186 |
| GO:0006508 | proteolysis and peptidolysis | 0.459068 | 1 | Thaps_bicluster_0186 |
| GO:0006355 | regulation of transcription, DNA-dependent | 0.481263 | 1 | Thaps_bicluster_0186 |

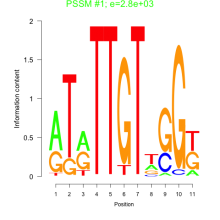
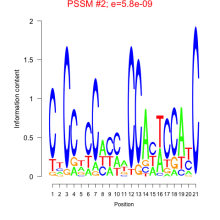
Comments