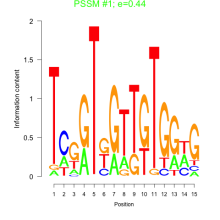
Thaps_bicluster_0189 Residual: 0.32
Thalassiosira pseudonana
| Title | Residual | Model version | Organism |
|---|---|---|---|
| Thaps_bicluster_0189 | 0.32 | Thalassiosira pseudonana |
Displaying 1 - 22 of 22
" class="views-fluidgrid-wrapper clear-block">
10796 hypothetical protein
11148 (Tp_HSF_3.8a) HSF_DNA-bind
21076 hypothetical protein
21602 NUC173 superfamily
21951 Sterol-sensing superfamily
22345 (PYK2) PLN02461
22721 hypothetical protein
24704 COG3899
25161 hypothetical protein
25417 hypothetical protein
25909 hypothetical protein
260975 (nhe3) Na_H_Exchanger
261601 PA14
262433 Pumilio superfamily
269238 (Tp_HSF_1.3e) HSF_DNA-bind superfamily
269900 (glsF) gltB
28239 PLN02272
3398 hypothetical protein
3478 hypothetical protein
40393 (PYK1) PLN02461
4972 hypothetical protein
9604 hypothetical protein
| GO ID | Go Term | p-value | q-value | Cluster |
|---|---|---|---|---|
| GO:0006096 | glycolysis | 0.0000865 | 0.237367 | Thaps_bicluster_0189 |
| GO:0006537 | glutamate biosynthesis | 0.00988969 | 1 | Thaps_bicluster_0189 |
| GO:0016071 | mRNA metabolism | 0.0221257 | 1 | Thaps_bicluster_0189 |
| GO:0006006 | glucose metabolism | 0.0245562 | 1 | Thaps_bicluster_0189 |
| GO:0006807 | nitrogen compound metabolism | 0.0269812 | 1 | Thaps_bicluster_0189 |
| GO:0015986 | ATP synthesis coupled proton transport | 0.108453 | 1 | Thaps_bicluster_0189 |
| GO:0006355 | regulation of transcription, DNA-dependent | 0.174355 | 1 | Thaps_bicluster_0189 |
| GO:0008152 | metabolism | 0.267706 | 1 | Thaps_bicluster_0189 |

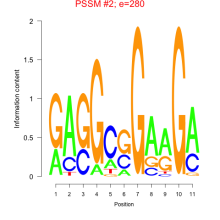
Comments