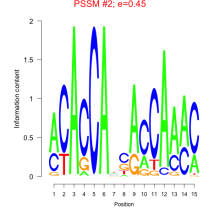
Thaps_bicluster_0196 Residual: 0.36
Thalassiosira pseudonana
| Title | Residual | Model version | Organism |
|---|---|---|---|
| Thaps_bicluster_0196 | 0.36 | Thalassiosira pseudonana |
Displaying 1 - 20 of 20
" class="views-fluidgrid-wrapper clear-block">
16947 DEXDc superfamily
20932 EEP superfamily
23331 RAP
23650 hypothetical protein
25247 Nudix_Hydrolase superfamily
25722 Beach superfamily
261769 GDA1_CD39 superfamily
261777 MIF4G
263669 UvrD
263987 hypothetical protein
264623 SpoVK
30980 EngA2
31259 HA
31764 DUF21
33670 RNRR2
33775 MetAP1
37156 PLN02369
42194 PTZ00184
4940 MiaA superfamily
6606 obgE
| GO ID | Go Term | p-value | q-value | Cluster |
|---|---|---|---|---|
| GO:0009186 | deoxyribonucleoside diphosphate metabolism | 0.00495457 | 1 | Thaps_bicluster_0196 |
| GO:0009165 | nucleotide biosynthesis | 0.00660132 | 1 | Thaps_bicluster_0196 |
| GO:0009116 | nucleoside metabolism | 0.00988766 | 1 | Thaps_bicluster_0196 |
| GO:0006298 | mismatch repair | 0.0261771 | 1 | Thaps_bicluster_0196 |
| GO:0008033 | tRNA processing | 0.0294066 | 1 | Thaps_bicluster_0196 |
| GO:0015031 | protein transport | 0.068993 | 1 | Thaps_bicluster_0196 |
| GO:0006118 | electron transport | 0.364179 | 1 | Thaps_bicluster_0196 |
| GO:0006508 | proteolysis and peptidolysis | 0.38827 | 1 | Thaps_bicluster_0196 |

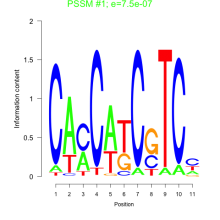
Comments