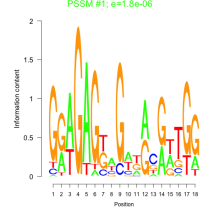
Thaps_bicluster_0197 Residual: 0.29
Thalassiosira pseudonana
| Title | Residual | Model version | Organism |
|---|---|---|---|
| Thaps_bicluster_0197 | 0.29 | Thalassiosira pseudonana |
Displaying 1 - 18 of 18
" class="views-fluidgrid-wrapper clear-block">
10417 hypothetical protein
11337 hypothetical protein
1136 hypothetical protein
17707 NADB_Rossmann superfamily
22652 UbiH
24343 hypothetical protein
25861 TPR
270331 hypothetical protein
33035 YGGT
4821 hypothetical protein
4980 TRAM_LAG1_CLN8
5045 hypothetical protein
5135 Rubis-subs-bind
5585 hypothetical protein
6504 CDP-OH_P_transf superfamily
6980 hypothetical protein
8720 NA
9513 hypothetical protein
| GO ID | Go Term | p-value | q-value | Cluster |
|---|---|---|---|---|
| GO:0006725 | aromatic compound metabolism | 0.026995 | 1 | Thaps_bicluster_0197 |
| GO:0006118 | electron transport | 0.287914 | 1 | Thaps_bicluster_0197 |
| GO:0008152 | metabolism | 0.410014 | 1 | Thaps_bicluster_0197 |
| GO:0008654 | phospholipid biosynthesis | 0.0172498 | 1 | Thaps_bicluster_0197 |
| GO:0018346 | protein amino acid prenylation | 0.00618938 | 1 | Thaps_bicluster_0197 |
| GO:0006457 | protein folding | 0.150875 | 1 | Thaps_bicluster_0197 |

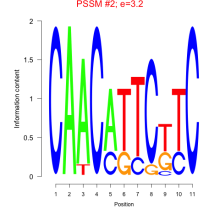
Comments