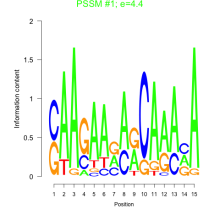
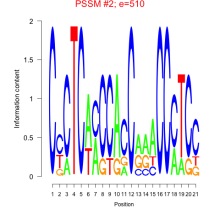
Thaps_bicluster_0202 Residual: 0.30
Thalassiosira pseudonana
| Title | Residual | Model version | Organism |
|---|---|---|---|
| Thaps_bicluster_0202 | 0.30 | Thalassiosira pseudonana |
Displaying 1 - 22 of 22
" class="views-fluidgrid-wrapper clear-block">
11154 hypothetical protein
13023 APP
13156 (ACS1) AFD_class_I superfamily
1912 2OG-FeII_Oxy_3 superfamily
21941 hypothetical protein
22096 SOUL
23958 Aldose_epim superfamily
24735 EMP70
25349 hypothetical protein
262207 Trp-synth-beta_II superfamily
264177 P4Hc
269232 EFh
3119 hypothetical protein
34876 (COP1) Coatomer_WDAD
37974 Sugar_tr
5094 hypothetical protein
6223 hypothetical protein
6243 Macro superfamily
6456 MhpC
6594 hypothetical protein
7874 hypothetical protein
8784 hypothetical protein
| GO ID | Go Term | p-value | q-value | Cluster |
|---|---|---|---|---|
| GO:0019538 | protein metabolism | 0.00645685 | 1 | Thaps_bicluster_0202 |
| GO:0006012 | galactose metabolism | 0.016026 | 1 | Thaps_bicluster_0202 |
| GO:0008643 | carbohydrate transport | 0.0370252 | 1 | Thaps_bicluster_0202 |
| GO:0006520 | amino acid metabolism | 0.0550594 | 1 | Thaps_bicluster_0202 |
| GO:0007186 | G-protein coupled receptor protein signaling pathway | 0.0550594 | 1 | Thaps_bicluster_0202 |
| GO:0006725 | aromatic compound metabolism | 0.0576102 | 1 | Thaps_bicluster_0202 |
| GO:0006810 | transport | 0.0852716 | 1 | Thaps_bicluster_0202 |
| GO:0006886 | intracellular protein transport | 0.135954 | 1 | Thaps_bicluster_0202 |
| GO:0008152 | metabolism | 0.300056 | 1 | Thaps_bicluster_0202 |
| GO:0006508 | proteolysis and peptidolysis | 0.550247 | 1 | Thaps_bicluster_0202 |

Comments