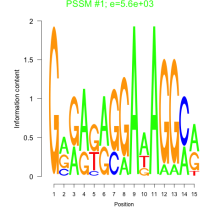
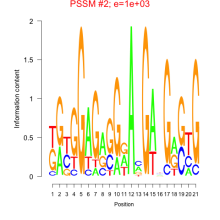
Thaps_bicluster_0205 Residual: 0.26
Thalassiosira pseudonana
| Title | Residual | Model version | Organism |
|---|---|---|---|
| Thaps_bicluster_0205 | 0.26 | Thalassiosira pseudonana |
Displaying 1 - 28 of 28
" class="views-fluidgrid-wrapper clear-block">
19541 (RL28) Ribosomal_L28e
20008 (RS26) PLN00186
22350 (RS21) Ribosomal_S21e
22535 (RS11) Ribosomal_S17 superfamily
25242 hypothetical protein
25949 (RL11A) PTZ00156
26250 (RL26) KOW_RPL26
26383 PTZ00255
26893 (RS18) PTZ00134
269086 (RS24) PTZ00071
27101 Ribosomal_L31e
28209 (RS23) Ribosomal_S23
30036 (RS27) PLN00209
31277 (RS30) Ribosomal_S30
31796 (RL39) Ribosomal_L39
32675 (RS15B) PTZ00096
32752 (RL24) Ribosomal_L24e_L24 superfamily
33977 (RL36) Ribosomal_L36e
34368 (RL37A) PTZ00073
39499 (RL14) Ribosomal_L14e
40312 (RS16) PLN00210
40387 (RL34) PTZ00074
42114 (RL35) Ribosomal_L29_HIP
42332 (RS29) Ribosomal_S14 superfamily
42457 (RS25) Ribosomal_S25
5259 (RL44) PTZ00157
6071 (RL22) Ribosomal_L22e
6363 (RL23a) PTZ00191
| GO ID | Go Term | p-value | q-value | Cluster |
|---|---|---|---|---|
| GO:0006412 | protein biosynthesis | 1.14e-39 | 3.16e-36 | Thaps_bicluster_0205 |

Comments