Thaps_bicluster_0208 Residual: 0.25
Thalassiosira pseudonana
| Title | Residual | Model version | Organism |
|---|---|---|---|
| Thaps_bicluster_0208 | 0.25 | Thalassiosira pseudonana |
Displaying 1 - 17 of 17
" class="views-fluidgrid-wrapper clear-block">
21667 (Lhcf7) fucoxanthin chlorophyll a/c light-harvesting protein, major type
2342 (Lhcr11) fucoxanthin chlorophyll a/c light-harvesting protein
24080 (Lhcr1) fucoxanthin chl a/c light-harvesting protein, lhcr type
25402 (Lhcf10) fucoxanthin chl a/c light-harvesting protein, major type
260392 (Lhcf2) fucoxanthin chlorophyll a/c protein 2
261232 URO-D
262032 hypothetical protein
270221 hypothetical protein
270233 hypothetical protein
270241 (Lhcf11) fucoxanthin-chlorophyll a-c binding protein, plastid precursor
270280 hypothetical protein
270316 hypothetical protein
32201 Cob-chelat-sub
35871 2-Hacid_dh_1
37615 NADB_Rossmann superfamily
4440 hypothetical protein
5240 ALAD_PBGS_aspartate_rich
| GO ID | Go Term | p-value | q-value | Cluster |
|---|---|---|---|---|
| GO:0015995 | chlorophyll biosynthesis | 0.00412967 | 1 | Thaps_bicluster_0208 |
| GO:0006783 | heme biosynthesis | 0.00412967 | 1 | Thaps_bicluster_0208 |
| GO:0006564 | L-serine biosynthesis | 0.00927255 | 1 | Thaps_bicluster_0208 |
| GO:0006779 | porphyrin biosynthesis | 0.0113237 | 1 | Thaps_bicluster_0208 |
| GO:0015979 | photosynthesis | 0.0133715 | 1 | Thaps_bicluster_0208 |

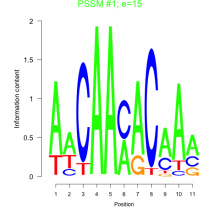

Comments