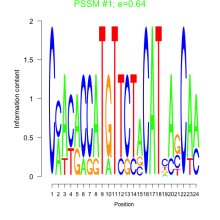
Thaps_bicluster_0211 Residual: 0.43
Thalassiosira pseudonana
| Title | Residual | Model version | Organism |
|---|---|---|---|
| Thaps_bicluster_0211 | 0.43 | Thalassiosira pseudonana |
Displaying 1 - 25 of 25
" class="views-fluidgrid-wrapper clear-block">
11018 hypothetical protein
11929 hypothetical protein
12122 hypothetical protein
1505 hypothetical protein
16503 AAA
21374 Lactonase
21554 hypothetical protein
24585 Syntaxin-6_N superfamily
24712 SrmB
24867 hypothetical protein
25267 hypothetical protein
25771 hypothetical protein
261566 Bud13
264821 PKc_like superfamily
269210 RsuA
30871 HAM1
37197 Pumilio superfamily
38225 MPN_eIF3f
39315 M20_dipept_like_CNDP
42515 DEADc
42543 2OG-FeII_Oxy_2
649 SrmB
8019 ovoA_Nterm
8235 hypothetical protein
9608 hypothetical protein
| GO ID | Go Term | p-value | q-value | Cluster |
|---|---|---|---|---|
| GO:0016071 | mRNA metabolism | 0.0129601 | 1 | Thaps_bicluster_0211 |
| GO:0006352 | transcription initiation | 0.0186738 | 1 | Thaps_bicluster_0211 |
| GO:0006508 | proteolysis and peptidolysis | 0.0608081 | 1 | Thaps_bicluster_0211 |
| GO:0006396 | RNA processing | 0.081025 | 1 | Thaps_bicluster_0211 |
| GO:0006468 | protein amino acid phosphorylation | 0.270534 | 1 | Thaps_bicluster_0211 |
| GO:0006355 | regulation of transcription, DNA-dependent | 0.368277 | 1 | Thaps_bicluster_0211 |


Comments