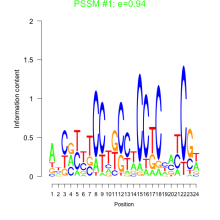
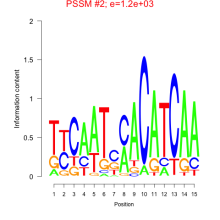
Thaps_bicluster_0217 Residual: 0.33
Thalassiosira pseudonana
| Title | Residual | Model version | Organism |
|---|---|---|---|
| Thaps_bicluster_0217 | 0.33 | Thalassiosira pseudonana |
Displaying 1 - 25 of 25
" class="views-fluidgrid-wrapper clear-block">
10740 hypothetical protein
11841 hypothetical protein
17623 SUMT
20827 hypothetical protein
20829 HPP
21143 hypothetical protein
21623 hypothetical protein
22251 hypothetical protein
22720 hypothetical protein
22731 hypothetical protein
24172 hypothetical protein
24341 DnaJ
25123 hypothetical protein
268279 hypothetical protein
269213 hypothetical protein
32758 Ntn_hydrolase superfamily
39516 PLN02951
4067 hypothetical protein
4919 hypothetical protein
5394 hypothetical protein
5424 hypothetical protein
6370 SpoU_methylase superfamily
6958 (Tp_HSF_4e) HSF_DNA-bind
8407 hypothetical protein
8521 PseudoU_synth superfamily
| GO ID | Go Term | p-value | q-value | Cluster |
|---|---|---|---|---|
| GO:0016567 | protein ubiquitination | 0.0108879 | 1 | Thaps_bicluster_0217 |
| GO:0006777 | Mo-molybdopterin cofactor biosynthesis | 0.0123468 | 1 | Thaps_bicluster_0217 |
| GO:0006779 | porphyrin biosynthesis | 0.0225308 | 1 | Thaps_bicluster_0217 |
| GO:0006511 | ubiquitin-dependent protein catabolism | 0.100662 | 1 | Thaps_bicluster_0217 |
| GO:0006396 | RNA processing | 0.113742 | 1 | Thaps_bicluster_0217 |
| GO:0006457 | protein folding | 0.238676 | 1 | Thaps_bicluster_0217 |
| GO:0006118 | electron transport | 0.432304 | 1 | Thaps_bicluster_0217 |
| GO:0006355 | regulation of transcription, DNA-dependent | 0.481263 | 1 | Thaps_bicluster_0217 |
| GO:0008152 | metabolism | 0.585137 | 1 | Thaps_bicluster_0217 |

Comments