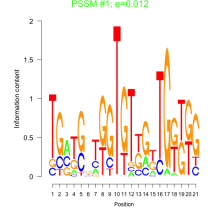
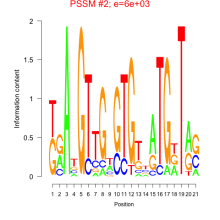
Thaps_bicluster_0222 Residual: 0.32
Thalassiosira pseudonana
| Title | Residual | Model version | Organism |
|---|---|---|---|
| Thaps_bicluster_0222 | 0.32 | Thalassiosira pseudonana |
Displaying 1 - 28 of 28
" class="views-fluidgrid-wrapper clear-block">
10434 hypothetical protein
11069 UAA
11186 hypothetical protein
1365 hypothetical protein
17008 RNase_HII_eukaryota_like
19793 PLN00157
20587 hypothetical protein
20776 hypothetical protein
21105 Acetyltransf_13
262697 hypothetical protein
264495 hypothetical protein
264676 S_TKc
268078 MFS_1
32223 Rab18
3280 COG0790
33109 QOR2
33578 Rab
3579 hypothetical protein
35968 RecA-like_NTPases superfamily
37328 PriL_PriS_Eukaryotic
4042 hypothetical protein
5009 hypothetical protein
5868 hypothetical protein
6652 hypothetical protein
7027 ArfGap superfamily
7090 hypothetical protein
7904 hypothetical protein
8486 hypothetical protein
| GO ID | Go Term | p-value | q-value | Cluster |
|---|---|---|---|---|
| GO:0007001 | chromosome organization and biogenesis (sensu Eukaryota) | 0.080847 | 1 | Thaps_bicluster_0222 |
| GO:0006334 | nucleosome assembly | 0.0861925 | 1 | Thaps_bicluster_0222 |
| GO:0006886 | intracellular protein transport | 0.145627 | 1 | Thaps_bicluster_0222 |
| GO:0006810 | transport | 0.419404 | 1 | Thaps_bicluster_0222 |
| GO:0006468 | protein amino acid phosphorylation | 0.468128 | 1 | Thaps_bicluster_0222 |
| GO:0006355 | regulation of transcription, DNA-dependent | 0.6012 | 1 | Thaps_bicluster_0222 |
| GO:0008152 | metabolism | 0.708368 | 1 | Thaps_bicluster_0222 |
| GO:0007264 | small GTPase mediated signal transduction | 0.00413978 | 1 | Thaps_bicluster_0222 |
| GO:0015031 | protein transport | 0.00656512 | 1 | Thaps_bicluster_0222 |
| GO:0006269 | DNA replication, synthesis of RNA primer | 0.00865975 | 1 | Thaps_bicluster_0222 |

Comments