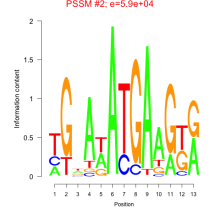
Thaps_bicluster_0225 Residual: 0.26
Thalassiosira pseudonana
| Title | Residual | Model version | Organism |
|---|---|---|---|
| Thaps_bicluster_0225 | 0.26 | Thalassiosira pseudonana |
Displaying 1 - 21 of 21
" class="views-fluidgrid-wrapper clear-block">
1254 Chalcone superfamily
1738 S14_ClpP_2
2078 hypothetical protein
21081 hypothetical protein
21821 hypothetical protein
2386 hypothetical protein
24700 hypothetical protein
261748 COG0523
262796 (ACP3) acpP
262849 Chromate_transp
270242 (PsbM) photosystem II reaction center M protein, plastid precursor
31673 PRK05481
32955 QOR2
33000 FTHFS
3353 FMN_dh
34030 (MAO1) PLN03129
34585 PLN02389
36462 (TPI3) TIM
4370 hypothetical protein
5576 DUF1499 superfamily
9485 mTERF superfamily
| GO ID | Go Term | p-value | q-value | Cluster |
|---|---|---|---|---|
| GO:0015703 | chromate transport | 0.00227414 | 1 | Thaps_bicluster_0225 |
| GO:0009102 | biotin biosynthesis | 0.00454357 | 1 | Thaps_bicluster_0225 |
| GO:0006108 | malate metabolism | 0.00680831 | 1 | Thaps_bicluster_0225 |
| GO:0009107 | lipoate biosynthesis | 0.00906837 | 1 | Thaps_bicluster_0225 |
| GO:0009396 | folic acid and derivative biosynthesis | 0.0158205 | 1 | Thaps_bicluster_0225 |
| GO:0006633 | fatty acid biosynthesis | 0.0424129 | 1 | Thaps_bicluster_0225 |
| GO:0006464 | protein modification | 0.126406 | 1 | Thaps_bicluster_0225 |
| GO:0006412 | protein biosynthesis | 0.319237 | 1 | Thaps_bicluster_0225 |
| GO:0006118 | electron transport | 0.463588 | 1 | Thaps_bicluster_0225 |
| GO:0006508 | proteolysis and peptidolysis | 0.491343 | 1 | Thaps_bicluster_0225 |

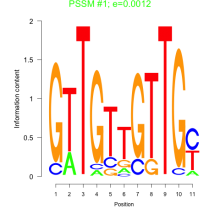
Comments