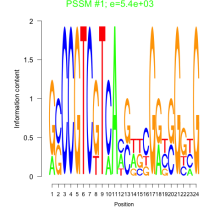
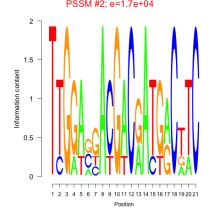
Thaps_bicluster_0227 Residual: 0.33
Thalassiosira pseudonana
| Title | Residual | Model version | Organism |
|---|---|---|---|
| Thaps_bicluster_0227 | 0.33 | Thalassiosira pseudonana |
Displaying 1 - 33 of 33
" class="views-fluidgrid-wrapper clear-block">
21871 (RSP1) PTZ00254
22476 (RL13e) Ribosomal_L13e superfamily
24423 (rpl23) PTZ00054
26046 (RS3A) Ribosomal_S3Ae
26063 WD40
26137 (RL9) PTZ00027
26221 (RS13) PTZ00072
26367 (RS15A) PTZ00158
264201 (RL8) PTZ00180
268651 (RS9) PLN00189
269148 PLN00116
269235 (RL3) PTZ00103
269779 (RS6) Ribosomal_S6e superfamily
269961 Ribosomal_L7Ae superfamily
27167 (RL7) Ribosomal_L7_archeal_euk
27292 (RS4) PLN00036
28049 PTZ00084
29007 (RL21) PLN00190
29217 PTZ00106
29825 (RS8) PTZ00148
29955 (RS5) PTZ00091
30154 (Rpl27) KOW_RPL27
33241 (RL17) Ribosomal_L22
37032 (RL6) KOW_RPL6
37628 Ribosomal_L7Ae
39282 (RL15) PTZ00026
39424 (RL12) PLN03072
39550 (RS7) Ribosomal_S7e
39735 (RL27A) Ribosomal_L18e superfamily
40329 (RL18a) RPL20A
41319 (RL18) Ribosomal_L18e superfamily
46 (RL13A) Ribosomal_L13
802 (RL5) PTZ00069
| GO ID | Go Term | p-value | q-value | Cluster |
|---|---|---|---|---|
| GO:0006412 | protein biosynthesis | 1.12104e-44 | 3.06996e-41 | Thaps_bicluster_0227 |
| GO:0042254 | ribosome biogenesis and assembly | 0.0468189 | 1 | Thaps_bicluster_0227 |
| GO:0006508 | proteolysis and peptidolysis | 0.869022 | 1 | Thaps_bicluster_0227 |

Comments