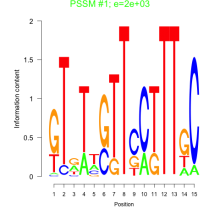
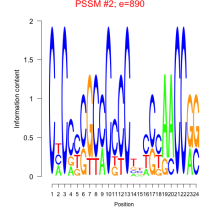
Thaps_bicluster_0228 Residual: 0.34
Thalassiosira pseudonana
| Title | Residual | Model version | Organism |
|---|---|---|---|
| Thaps_bicluster_0228 | 0.34 | Thalassiosira pseudonana |
Displaying 1 - 23 of 23
" class="views-fluidgrid-wrapper clear-block">
10039 hypothetical protein
11061 hypothetical protein
11100 hypothetical protein
14492 S_TKc
18624 Mpv17_PMP22
20186 DnaJ
20952 MscS
21645 hypothetical protein
23503 ANK
23642 hypothetical protein
23692 hypothetical protein
24003 (Tp_HSF_2.5h) HSF_DNA-bind superfamily
24923 hypothetical protein
25122 RING
25261 hypothetical protein
25292 hypothetical protein
25306 hypothetical protein
25452 hypothetical protein
3196 hypothetical protein
3839 hypothetical protein
4411 hypothetical protein
7135 hypothetical protein
9958 hypothetical protein
| GO ID | Go Term | p-value | q-value | Cluster |
|---|---|---|---|---|
| GO:0016567 | protein ubiquitination | 0.0637659 | 1 | Thaps_bicluster_0228 |
| GO:0006457 | protein folding | 0.103279 | 1 | Thaps_bicluster_0228 |
| GO:0006468 | protein amino acid phosphorylation | 0.164893 | 1 | Thaps_bicluster_0228 |
| GO:0006355 | regulation of transcription, DNA-dependent | 0.230778 | 1 | Thaps_bicluster_0228 |

Comments