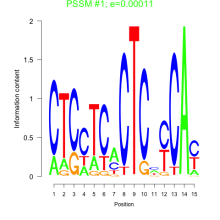
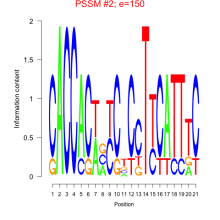
Thaps_bicluster_0230 Residual: 0.24
Thalassiosira pseudonana
| Title | Residual | Model version | Organism |
|---|---|---|---|
| Thaps_bicluster_0230 | 0.24 | Thalassiosira pseudonana |
Displaying 1 - 15 of 15
" class="views-fluidgrid-wrapper clear-block">
20814 hypothetical protein
22036 ElaC
22628 hypothetical protein
24217 hypothetical protein
24715 WD40 superfamily
24878 COG0412
25139 RRN3 superfamily
262041 MIF4G
268145 PLN00040
268596 CBF
29705 18S_RNA_Rcl1p
34413 RRM_ABT1_like
40265 PTZ00448
42152 WD40
4372 hypothetical protein
| GO ID | Go Term | p-value | q-value | Cluster |
|---|---|---|---|---|
| GO:0007186 | G-protein coupled receptor protein signaling pathway | 0.00434153 | 1 | Thaps_bicluster_0230 |

Comments