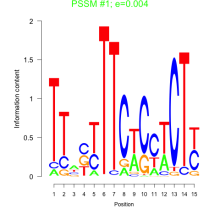
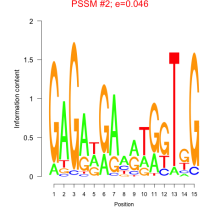
Thaps_bicluster_0251 Residual: 0.43
Thalassiosira pseudonana
| Title | Residual | Model version | Organism |
|---|---|---|---|
| Thaps_bicluster_0251 | 0.43 | Thalassiosira pseudonana |
Displaying 1 - 24 of 24
" class="views-fluidgrid-wrapper clear-block">
11508 Prefoldin_beta
14708 DRE_TIM_metallolyase superfamily
22426 hypothetical protein
22929 hypothetical protein
24164 hypothetical protein
24345 Cupin_8 superfamily
2523 hypothetical protein
2605 hypothetical protein
261694 MPP_PAPs
262538 hypothetical protein
262707 Cupin_8 superfamily
263386 GDA1_CD39 superfamily
269717 NBD_sugar-kinase_HSP70_actin superfamily
269828 hypothetical protein
2938 hypothetical protein
36650 hypothetical protein
5332 hypothetical protein
5711 hypothetical protein
6376 hypothetical protein
8438 hypothetical protein
9127 Smc
9292 hypothetical protein
9384 hypothetical protein
9476 hypothetical protein
| GO ID | Go Term | p-value | q-value | Cluster |
|---|---|---|---|---|
| GO:0030163 | protein catabolism | 0.0107105 | 1 | Thaps_bicluster_0251 |
| GO:0015986 | ATP synthesis coupled proton transport | 0.0375124 | 1 | Thaps_bicluster_0251 |
| GO:0005975 | carbohydrate metabolism | 0.0606126 | 1 | Thaps_bicluster_0251 |
| GO:0016567 | protein ubiquitination | 0.0637659 | 1 | Thaps_bicluster_0251 |

Comments