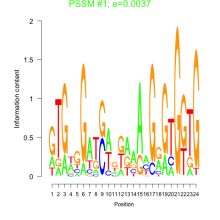
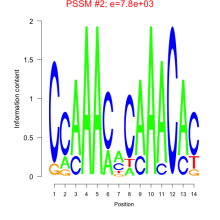
Thaps_bicluster_0259 Residual: 0.35
Thalassiosira pseudonana
| Title | Residual | Model version | Organism |
|---|---|---|---|
| Thaps_bicluster_0259 | 0.35 | Thalassiosira pseudonana |
Displaying 1 - 24 of 24
" class="views-fluidgrid-wrapper clear-block">
10252 hypothetical protein
11147 HP_PGM_like
11172 hypothetical protein
21908 hypothetical protein
21925 NADB_Rossmann superfamily
2200 hypothetical protein
22261 hypothetical protein
22854 PTS_2-RNA
24879 hypothetical protein
25138 Methyltransf_32
25359 hypothetical protein
25730 Rdx superfamily
25800 hypothetical protein
268449 Thioredoxin_like superfamily
270004 hypothetical protein
2890 hypothetical protein
32766 COG1936
38646 TRX_family
38679 UBQ superfamily
39698 Zn-ribbon_RPA12
42412 RNAP_I_II_AC40
5806 TIM_phosphate_binding superfamily
6380 hypothetical protein
7031 CIA30 superfamily
| GO ID | Go Term | p-value | q-value | Cluster |
|---|---|---|---|---|
| GO:0006388 | tRNA splicing | 0.00186066 | 1 | Thaps_bicluster_0259 |
| GO:0006354 | RNA elongation | 0.00742418 | 1 | Thaps_bicluster_0259 |
| GO:0006568 | tryptophan metabolism | 0.0111179 | 1 | Thaps_bicluster_0259 |
| GO:0006350 | transcription | 0.0544987 | 1 | Thaps_bicluster_0259 |
| GO:0006118 | electron transport | 0.0839917 | 1 | Thaps_bicluster_0259 |
| GO:0006464 | protein modification | 0.104655 | 1 | Thaps_bicluster_0259 |
| GO:0006355 | regulation of transcription, DNA-dependent | 0.446038 | 1 | Thaps_bicluster_0259 |
| GO:0008152 | metabolism | 0.546944 | 1 | Thaps_bicluster_0259 |

Comments