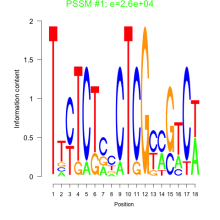
Thaps_bicluster_0267 Residual: 0.26
Thalassiosira pseudonana
| Title | Residual | Model version | Organism |
|---|---|---|---|
| Thaps_bicluster_0267 | 0.26 | Thalassiosira pseudonana |
Displaying 1 - 16 of 16
" class="views-fluidgrid-wrapper clear-block">
11041 hypothetical protein
11557 PhyH superfamily
11707 VDE superfamily
11760 hypothetical protein
1464 hypothetical protein
1724 hypothetical protein
1786 hypothetical protein
21114 Vac14_Fab1_bd
22689 Chalcone superfamily
2767 AarF
3622 CDP-ME_synthetase
36431 cyclophilin_TLP40_like
3884 hypothetical protein
4040 hypothetical protein
6759 hypothetical protein
8085 hypothetical protein
| GO ID | Go Term | p-value | q-value | Cluster |
|---|---|---|---|---|
| GO:0008299 | isoprenoid biosynthesis | 0.00680831 | 1 | Thaps_bicluster_0267 |
| GO:0006468 | protein amino acid phosphorylation | 0.1264 | 1 | Thaps_bicluster_0267 |
| GO:0006457 | protein folding | 0.0784967 | 1 | Thaps_bicluster_0267 |

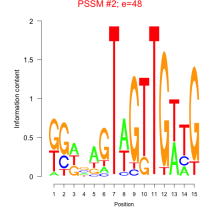
Comments